1. Introduction
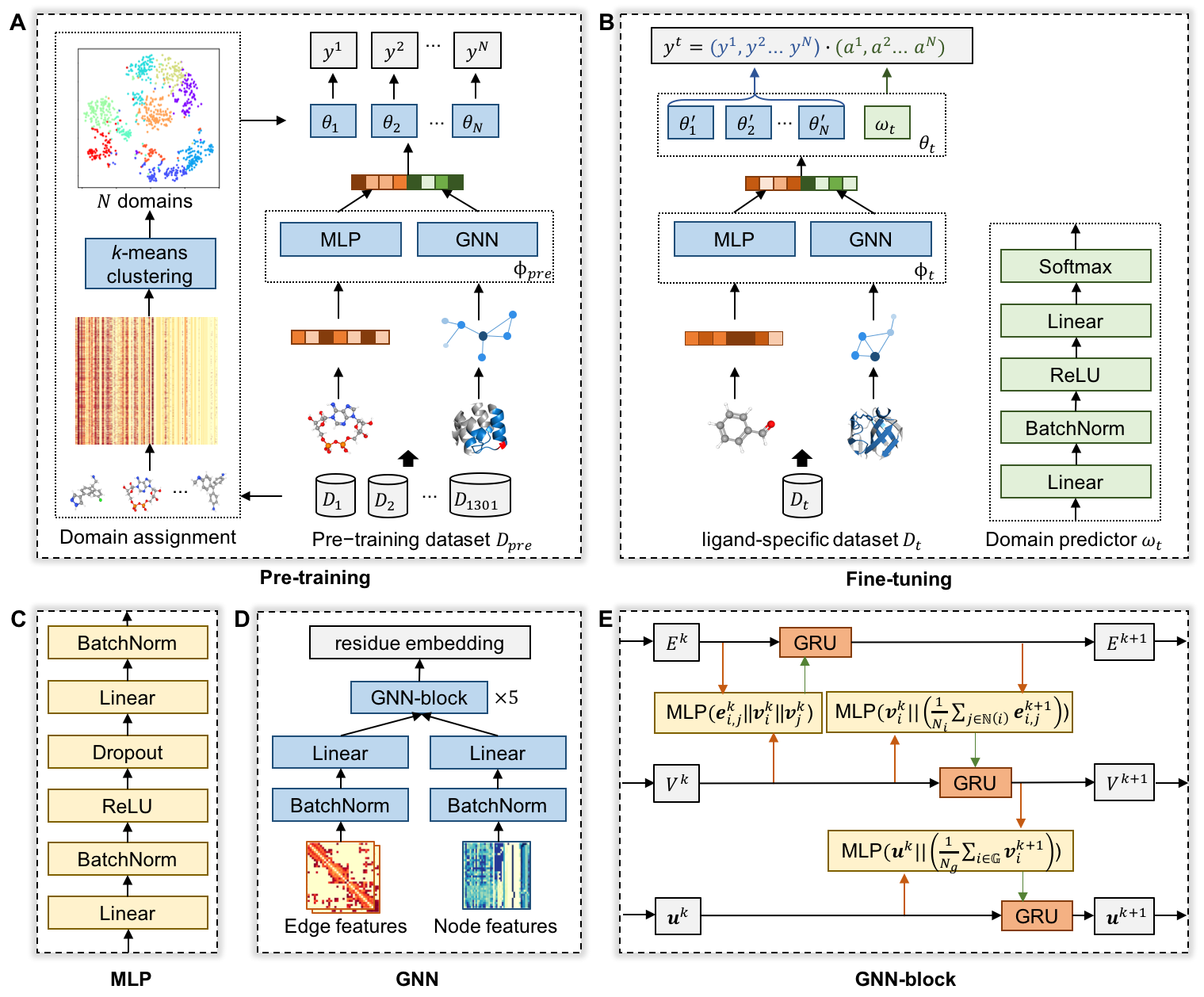
LigBind is a novel domain adaption-based framework with graph-level pre-training to predict the ligand-specific binding residues for over 1000 ligands from protein structures. LigBind consists of pre-training and fine-tuning phases: 1) pre-training learns relation-aware classifiers to distinguish the shared binding patterns of similar ligands from dissimilar ligands and alleviates the bias caused by the class imbalance; and 2) fine-tuning with ligand-specific data integrates the binding diversity into the predictors. We construct a pre-training dataset of 1301 ligands for model pre-training and 1159 ligand-specific benchmark datasets for constructing the ligand-specific predictors. The framework of LigBind and its backbone GNN is shown in Figure 1.
2. Input
First, for predicting ligand-binding residues, please input the protein chain structure (in PDB format)
and 1-character chain ID (chain ID is case sensitive. If your query chain doesn't contain chain ID, just leave the chain ID box blank).
There are three methods to choose from:
Method 1. LigBind. If the target ligand is included in 1159 ligands, you can select the ligand type and use ligand-specific LigBind for prediction.
You can search ligand ID by ligand name from ligands information.
Method 2. LigBind-G. If the target ligand isn't included in 1159 ligands, you can input the ligand SMILES and use ligand-general LigBind-G for prediction.
Method 3. LigBind-G. If you want to predict general ligand-binding residues without ligand information, you can choose this method.
For a query protein, it takes about 5 minutes for prediction and we will send the results to your email when the job is finished.
3. Output
We will send the results to your email when the job is finished.
Results will be shown in the result page (example) when the job is finished. Binding residues are marked as red in the sequence extracted from the PDB file.
The 3D structure of query protein and binding residues are also shown in the JSmol. In addition, results can be downloaded by clicking "Download results".
