1. Introduction
GraphBind is a webserver designed for structure-based nucleic acid- and small ligand-binding residues prediction.
GraphBind consists of two modules:
1) Constructing graphs based on structure, which integrates the local neighborhood around residues to construct
graphs. Figure 1 shows the flowchart of constructing a graph based on structure context in GraphBind.
2) The hierarchical graph neural networks (HGNNs), which progressively updates the edge features, node features
and graph features, and further learns high-level features for classifying the binding residues.
Figure 2 and Figure 3 show the diagram of the hierarchical graph neural network and GNN-block, respectively.
Besides, we transfer the proposed HGNNs to GraphBind-G, a ligand generic model that is trained by proteins bound to hundreds of ligand types.
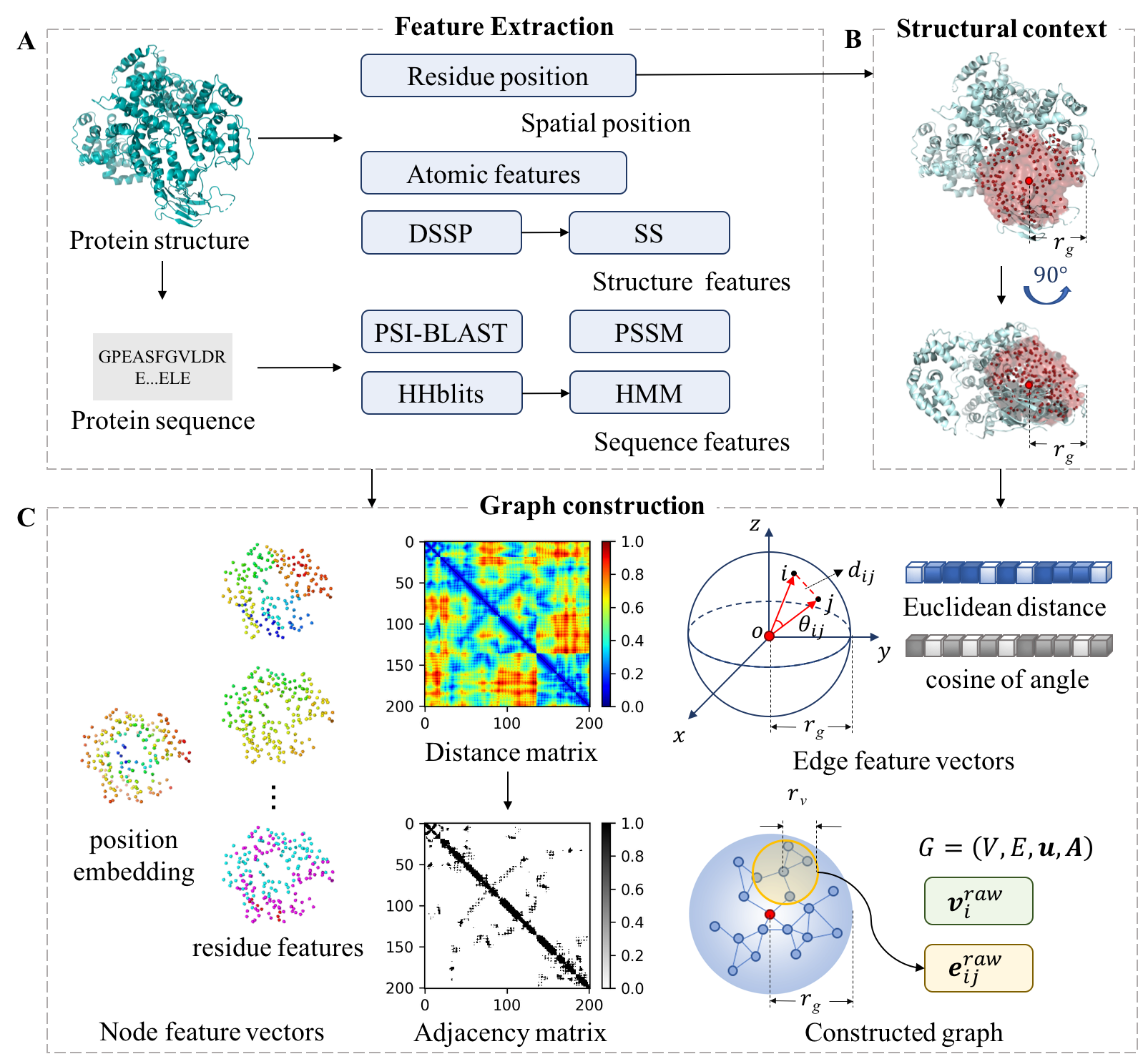
Figure 1.The flowchart of constructing a graph based on structure context in GraphBind.
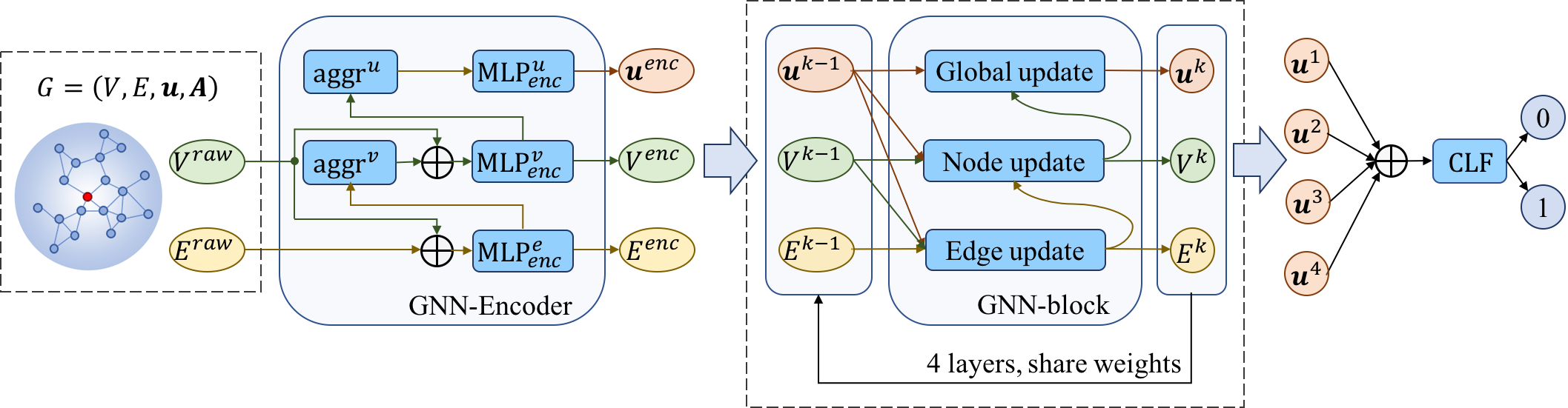
Figure 2.The diagram of the hierarchical graph neural network.
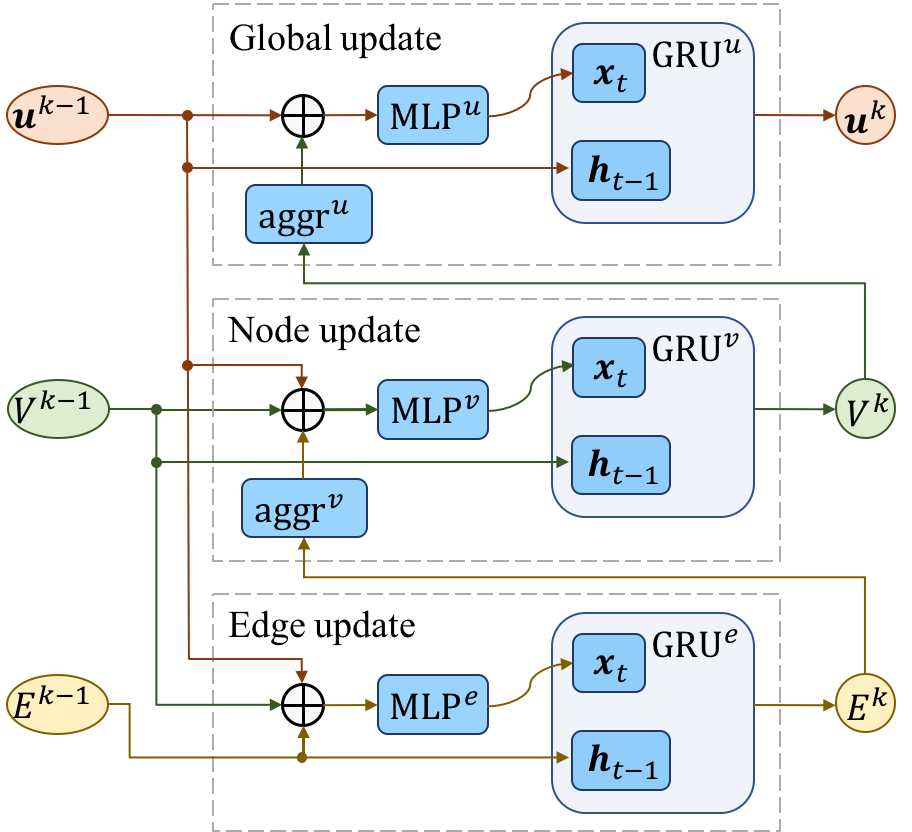
Figure 3.GNN-block consists of sequential edge update, node update and global update.
2. Inputs
GraphBind only supports processing one sequence at a time. Please input the protein chain structure
(in PDB format) and
1-character chain ID (chain ID is case sensitive, if your query chain doesn't contain chain ID,
just leave the chain ID box blank). There are two functions to choose from.
You can choose the ligand-specific GraphBind and the ligand types to predict ligand binding residues for the desired ligands.
Or you can use the ligand-general GraphBind-G to predict the binding residues for a general ligands.
Finally, you can input your Email address for receiving results.
Running time is related to the length of query protein chain and the condition of our server.
For example, the chain of length 100, 500, and 1000 takes approximately 10 minutes, 30 minutes, and an hour to finish, respectively.
Therefore, We do not recommend you to submit a very long protein chain.
3. Outputs
We will send the results to your email when the job is finished.
Results will be shown in the result page (example) when the job is finished.
Binding residues are marked as red in the sequence extracted from the PDB file.
The 3D structure of query protein and binding residues are also shown in the JSmol.
In addition, results can be downloaded by clicking "Download results".
This results can be saved for two weeks.
