
|
Enhancing Membrane Protein Subcellular Localization Prediction by Parallel Fusion of Multi-View Features
Introduction Membrane proteins are encoded by ~30% in the genome and function importantly in the living organisms. Previous studies have revealed that membrane proteins' structures and functions show obvious cell organelle-specific properties. Hence, it is highly desired to predict membrane protein's subcellular location from the primary sequence considering the extreme difficulties of membrane protein wet-lab studies. Although many models have been developed for predicting protein subcellular locations, very few are specific to membrane proteins. Existing prediction approaches were constructed based on statistical machine learning algorithms with serial combination of multi-view features, i.e., different feature vectors are simply serially combined to form a super feature vector. However, such simple combination of features will simultaneously increase the information redundancy that could, in turn, deteriorate the final prediction accuracy. That's why it was often found that prediction success rates in the serial super space were even lower than those in a single-view space. The purpose of this paper is investigation of a proper method for fusing multiple multi-view protein sequential features for subcellular location predictions. Instead of serial strategy, we propose a novel parallel framework for fusing multiple membrane protein multi-view attributes that will represent protein samples in complex spaces. We also proposed generalized principle component analysis (GPCA) for feature reduction purpose in the complex geometry. All the experimental results through different machine learning algorithms on benchmark membrane protein subcellular localization dataset demonstrate that the newly proposed parallel strategy outperforms the traditional serial approach. We also demonstrate the efficacy of the parallel strategy on a soluble protein subcellular localization dataset indicating the parallel technique is flexible to suite for other computational biology problems. Flowchart of Parallel Fusion of Multi-View Features Figure 1 shows the flowchart of parallel fusion of multi-view features and click here to download the whole software package and the benchmark datasets 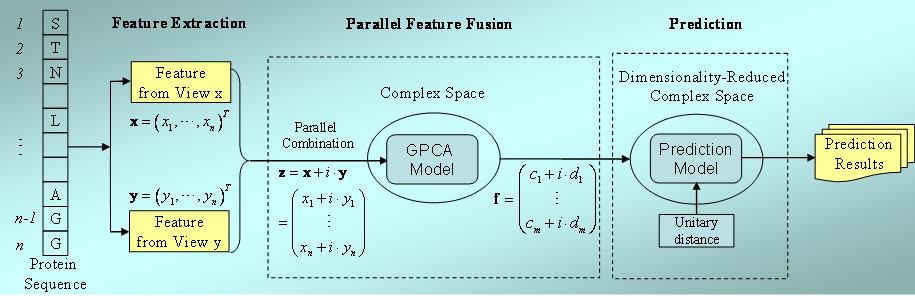 Figure 1. Flowchart of parallel fusion of multi-view features. Link MemBrain: Improving the Accuracy of Predicting Transmembrane Helices. SOMRuler: A Novel Interpretable Transmembrane Helices Predictor. Reference Dong-Jun Yu, Hong-Bin Shen, Xiao-Wei Wu, Jian Yang, Zhen-Min Tang, Yong Qi, and Jing-Yu Yang: Enhancing Membrane Protein Subcellular Localization Prediction by Parallel Fusion of Multi-View Features, IEEE Transactions on NanoBioscience, 2012, 11: 375-385. |