1. Introduction
Great improvement has been brought to protein tertiary structure prediction through deep learning.
It is important but very challenging to accurately rank and score decoy structures predicted by different models.
CASP14 results show that existing quality assessment (QA) approaches lag behind the development of protein structure prediction,
where almost all existing QA models degrade in accuracy when the target is a decoy of high quality, such as close to the native configuration.
How to give an accurate assessment of high-accuracy decoys is particularly useful with the available of accurate structure prediction methods.
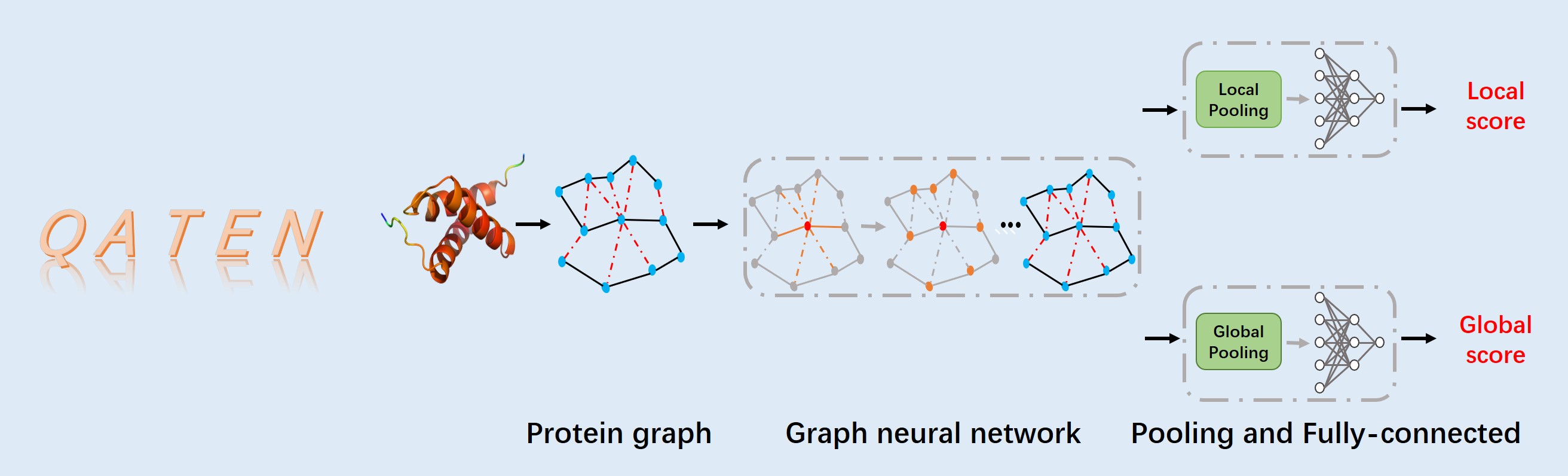
In this paper, we propose a fast and effective single-model QA method, QATEN,
which can evaluate decoys only by their topological characteristics and atomic types.
Our model uses graph neural networks and attention mechanisms to evaluate global and amino acid level scores,
and uses specific loss functions to constrain the network to focus more on high-precision decoys and high-precision protein domains.
On the CASP14 evaluation decoys, QATEN performs better than other QA models under all correlation coefficients when targeting average LDDT.
While it performs comparably to DeepAccNet when targeting GDT-TS.
Besides, QATEN shows state-of-the-art performance when considering only high-accuracy decoys.
Compared to the embedded evaluation modules of predicted Alpha-C RMSD in RosettaFold and predicted LDDT in AlphaFold2,
QATEN is complementary and capable of achieving better evaluation on some decoy structures generated by AlphaFold2 and RosettaFold themselves.
These results suggest that the new QATEN approach can be used as a reliable independent assessment algorithm for high-accuracy protein structure decoys.
Please notice that QATEN is only free for academic use.
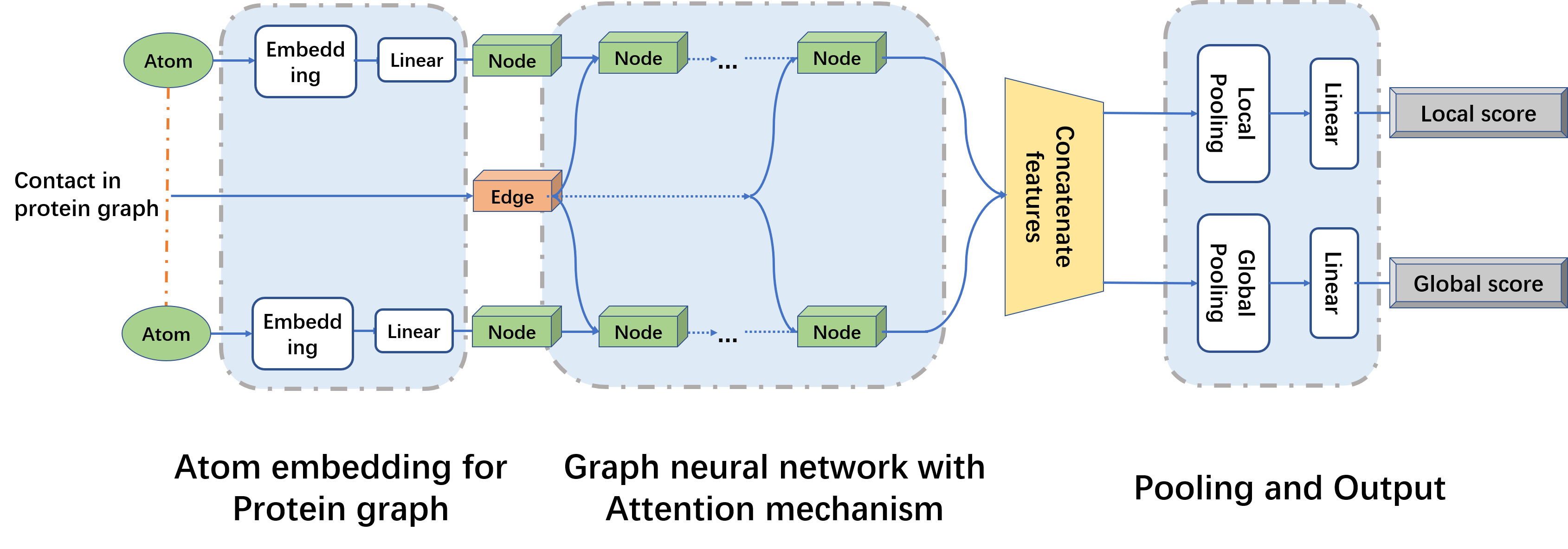
Figure 1.The flowchart of QATEN which mainly includes three steps.
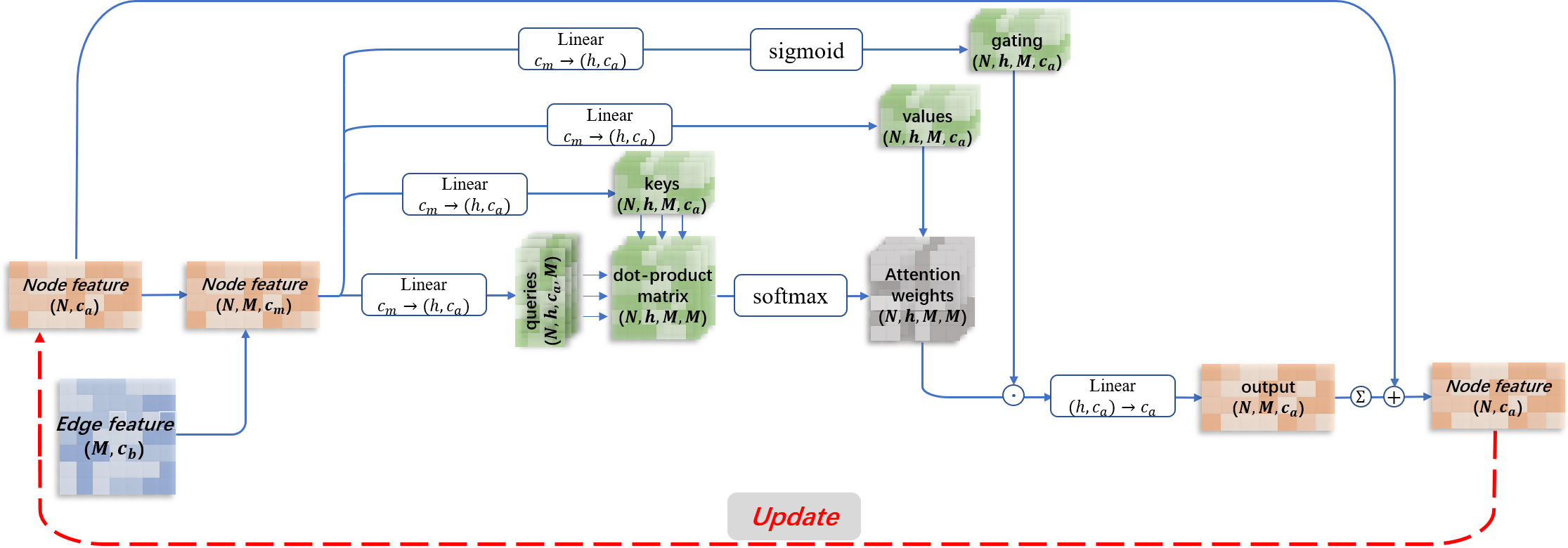
Figure 2.The attention module of graph neural networks in QATEN.
2. Inputs
QATEN online only supports processing one decoy at a time. Please input the protein decoy structure
(in PDB format).
You can also download the source code and run QATEN locally
(Source code). Please note that QATEN will only extract and predict chain A.
There are two results (global score and local scores) to be predicted, you can input your Email address for receiving results.
Running time is related to the length of query protein chain and the condition of our server.
For example, the chain of length 100, 500, and 1000 takes approximately 20 seconds, 1 minute, and 3 minutes to finish, respectively.
Therefore, We do not recommend you to submit a very long protein chain.
3. Outputs
We will send the results to your email when the job is finished (optional).
Results will be shown in the result page (example) when the job is finished.
We color each amino acid, using a bwr gradient, according to the "local scores" value.

(red = hot = high confidenc)
In addition, value results can be downloaded by clicking "Download results". This results can be saved for two weeks.