|
MemBrain Tutorial
|
| 1. Introduction | |
| MemBrain is a web server developed for transmembrane protein structure prediction. To date, it contains two main prediction functions, i.e., transmembrane helix (TMH) prediction and TMH-TMH residue contact prediction. | |
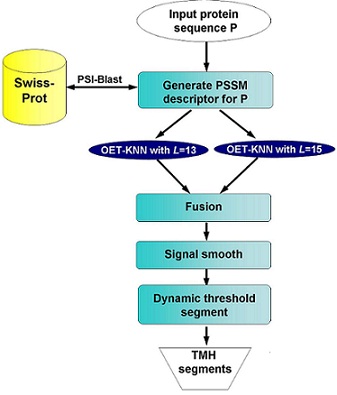 Figure 1. Flowchart of TMH prediction in MemBrain. |
1.1 TMH prediction Prediction of TMHs in alpha-helical membrane proteins provides valuable information about the protein topology when the high resolution structures are not available. To improve the accuracy of TMH detection, we developed a machine learning-based predictor, which integrates a number of modern bioinformatics approaches including sequence representation by multiple sequence alignment matrix, the optimized evidence-theoretic k-nearest neighbor prediction algorithm, fusion of multiple prediction window sizes, and classification by dynamic threshold. The result demonstrates the improvement of predicting the ends of TMHs and TMHs that are shorter than 15 residues. It also has the capability to detect N-terminal signal peptides. Figure 1 illustrates the flowchart of TMH prediction. |
|
1.2 TMH-TMH residue contact prediction Prediction of TMH-TMH residue contacts can provide crucial constraints for accurately constructing 3D structures of membrane proteins. For TMH-TMH residue contact prediction, TMH locations are derived from TMH prediction embeded in MemBrain. Recently, we developed a novel TMH-TMH contact map predictor for membrane proteins directly from the primary sequence, which is a new component of MemBrain protocol. It was constructed from the combination of statistical machine learning algorithms and biological evolution analysis from multiple sequence alignments as shown in Figure 2. The machine learning-based prediction engine was trained by applying multiple algorithms on multiple random under-samplings so that strong diversities can be generated via different learning methods in various spaces. The biological evolution analysis from multiple sequence alignments was done by PSICOV algorithm. MemBrain is a useful sequence-based analysis tool for functional and structural characterization of helical membrane proteins. |
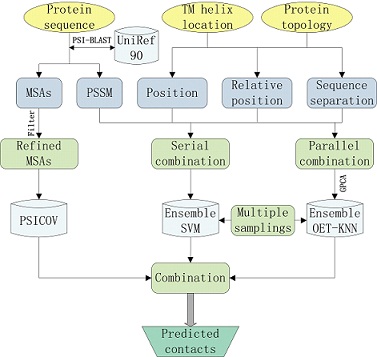 Figure 2. Flowchart of TMH-TMH residue contact prediction in MemBrain. |
|
1.3 TMP topology prediction (new model) The information of TMP structures is critically important to reveal their complex functions and for aiding the drug design. Prediction of TMP topology can be separated into two sub-tasks: (1) TMH prediction, and (2) orientation determination. For the first sub-task, a fusion prediction engine of multi-scale deep learning models is constructed, where the small-scale and large-scale models are residue-based and entire-sequence-based residual neural networks, respectively. Then, dynamic threshold decision strategy is designed to solve the over- and under- TMH splitting problem. For the second one, a support vector machine (SVM) model with new Max-Min score assignment approach is employed to determine the inside/outside position of each loop region.Amphipathic helix (AH) features the segregation of polar and nonpolar residues and plays important roles in many membrane-associated biological processes through interacting with both the lipid and the soluble phases. A new deep learning-based AH prediction pipeline is developed, which is composed of a residual neural network and uneven-thresholds decision algorithm. |
|
|
1.4 TMH-TMH residue contact prediction (deep learning model) The new MemBrain is a hierarchical two-stage residue contact predictor. For the first stage, it is a conventional two-hidden-layer perceptron. 1084-dimensional sequence-based features are fed into this neural network with 150 units for each of the two hidden layers. The single output indicates the contact potential of given residue pair. The second stage is the fusion of three powerful CNNs, which have one, two and three convolution layers respectively. On the top of each CNN, a fully connected layer with 150 hidden units is used to predict the final contact probability. Figure 3 illustrates the flow chart of MemBrain protocol. |
|
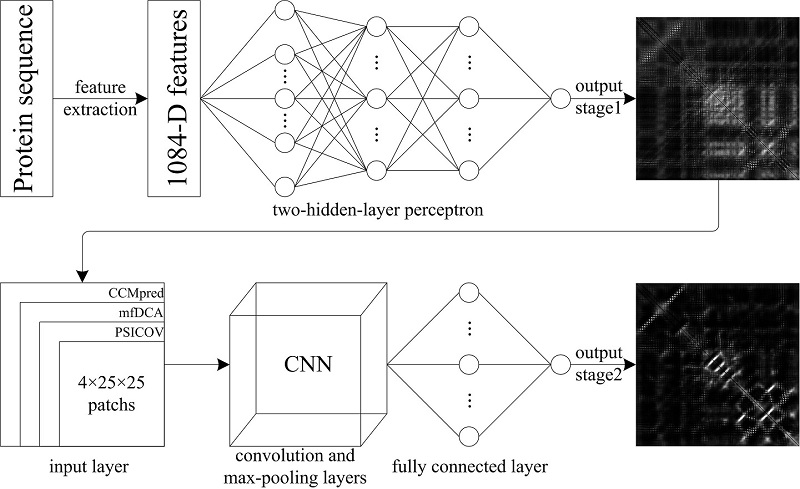 Figure 3. Flowchart of TMH-TMH residue contact prediction in new MemBrain. |
|
|
1.5 rASA prediction Prediction of rASA in alpha-helical transmembrane proteins provides the relative positions of the residues which is helpful to 3D structure prediction. To improve the performance of rASA prediction, We present a novel sequence-based method (MemBrain-Rasa) to predict relative solvent accessibility surface area from primary sequence. The MemBrain-Rasa features by a newly developed segment structural similarity-based prediction engine, which is further combined with the machine learning engine. We locally constructed a comprehensive database of residue relative solvent accessibility surface area, which is used to be searched for segments that are expected to be structural similar to the segments on the query sequence. The segment structural similarity-based prediction is then fused with the support vector regression outputs using a designed knowledge rule. |
|
|
1.6 3D structure model prediction MemBrain also provides the 3D structure model prediction function of membrane proteins. It is an ab initio prediction model. We use TMP topology prediction to obtain the secondary stucture information, use ResNet network to predict the residue contact. Under the guidance of this knowledge, We make tens of millions of fragment substitutions to get the final model.MemBrain also allows users to add constraint information about residue contact, so as to further improve the accuracy of prediction. It should be noted that the 3D structure model prediction function of MemBrain is only appropriate to those membrane proteins with transmembrane helix as the main part, and the accuracy will be reduced when there are many outer membrane parts. |
|
|
|
| 2. Inputs | |
|
(A). Protein sequence Input the protein sequence into the input box without ID, the sequence should contain more than 30 aa but not include invalid character, as shown by clicking Example hyperlink. | |
|
(B). Prediction function If you select "α-TMP Topology prediction using deep learning", MemBrain will predict the TMP topology according to your selection of "Transmembrane helix" and "Amphipathic helix". If you select "TMH prediction", MemBrain will only predict TMHs. If you select "TMH-TMH residue contact prediction", MemBrain will predict TMHs firstly, and then predict TMH-TMH residue contacts. If you select "Rasa prediction", MemBrain will predict real-value relative accessible surface area (rASA) for each amino acid. | |
|
(C). N-terminal signal peptide information MemBrain has the capability to detect N-terminal signal peptides. If you select "I know there is NO N-terminal signal peptide", MemBrain will not detect the N-terminal signal peptide (default). If you select "I do NOT know whether there is signal peptide in the N-terminal or not", MemBrain will apply "Signal-3L" predictor to automatically identify the N-terminal signal peptide. You must select the species of the query sequence below in this case. Note: N-terminal signal peptide information does not affect the prediction of TMH-TMH residue contacts. | |
|
(D). Email address You should input your email address to receive an email notification of your prediction results. | |
| 3. Outputs | |
|
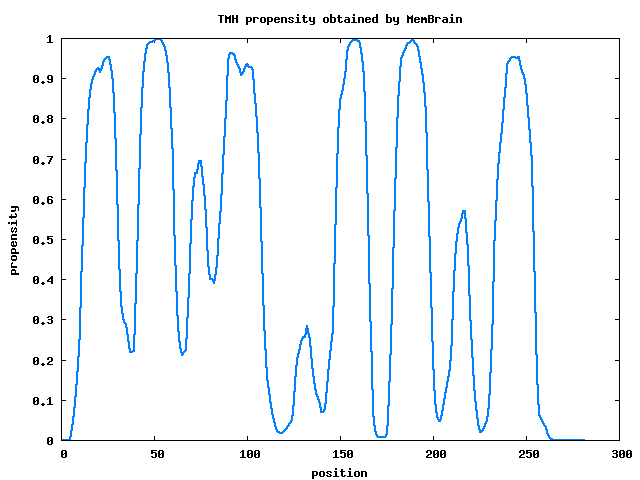
Case 1. TMH prediction The detailed text description of prediction results will be sent to your emaill, include N-terminal signal peptide information, predicted TMHs, and TMH propensities for each residue. The picture description of TMH propensities for each residue is shown in Figure 4.
| |
|
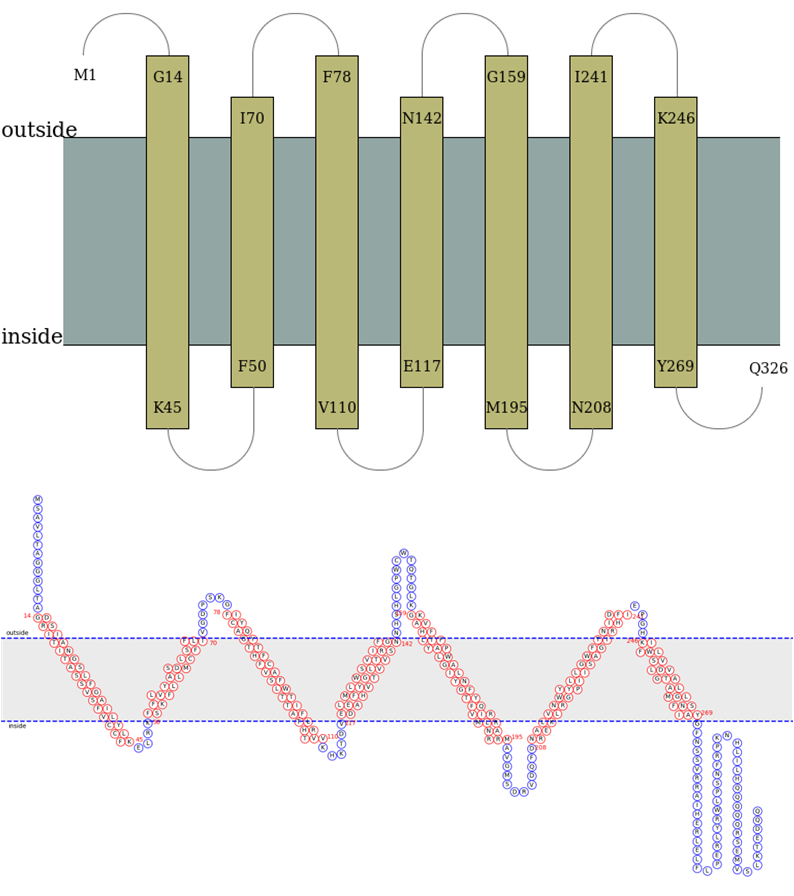
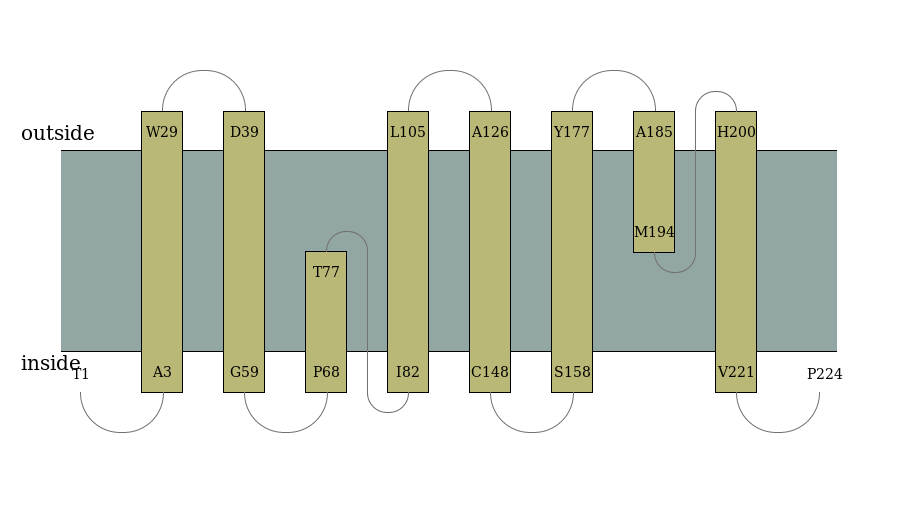
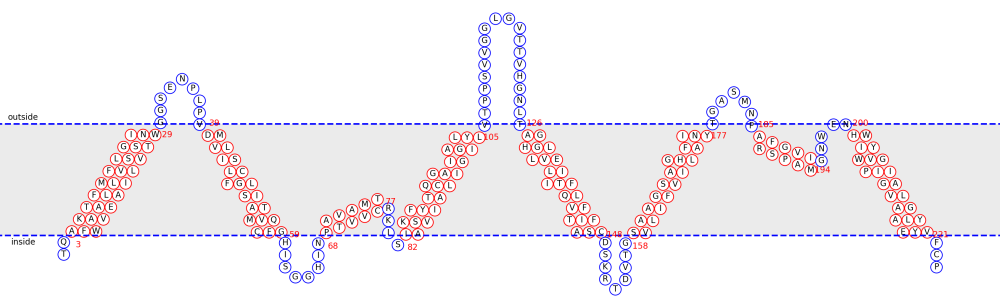
Case 2. α-TMP topology prediction The detailed text description of prediction results will be sent to your emaill, along with the THM propensities figure and topology illustration figure.[Example] The picture description of TMP topology is shown in Figure 5.
| |
|
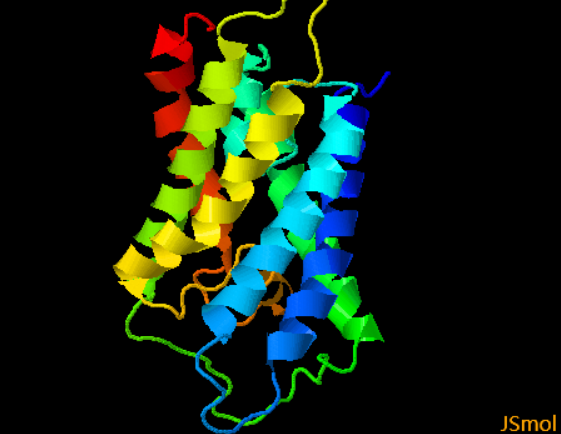
Case 3. TMH-TMH residue contact prediction Other than the outputs for TMH prediction described above, MemBrain also outputs detailed prediction results for TMH-TMH residue contact prediction, include predicted TMHs, predicted contact map and detailed information. [Example] Case 4. Rasa prediction The detailed text description of rASA predictions will be sent to your emaill. Most of the rASA values lie in the range [0, 100]. If rASA is greater than 100%, probably because the residue is next to a chain break or there have some unusual bond angles, bond lengths and distorted geometry in real proteins. If rASA is lower than 0, probably because the machine learning engine generates predictions lower than 0. Case 5. 3D structure model prediction The output part mainly contains a pdb file, which will be displayed on the web page, and the intermediate results of the prediction process, including the topology, will also be given.
| |