Background
The advent of highly accurate protein structure prediction methods has fueled an exponential expansion of the protein structure database. Consequently, there is a rising demand for rapid and precise structural homolog search. Traditional alignment-based methods are dedicated to precise comparisons between pairs, exhibiting high accuracy. However, their sluggish processing speed is no longer adequate for managing the current massive volume of data. In response to this challenge, we propose a novel deep-learning approach FoldExplorer.
The workflow is as following:
 Step 1: Input you query structure, which should be PDB or mmCIF format.
Step 1: Input you query structure, which should be PDB or mmCIF format.
Step 2: Select a database you want to search against.
Step 3: Click the "Submit" button and wait for a minute.
Then you can click the link and go to the result page.
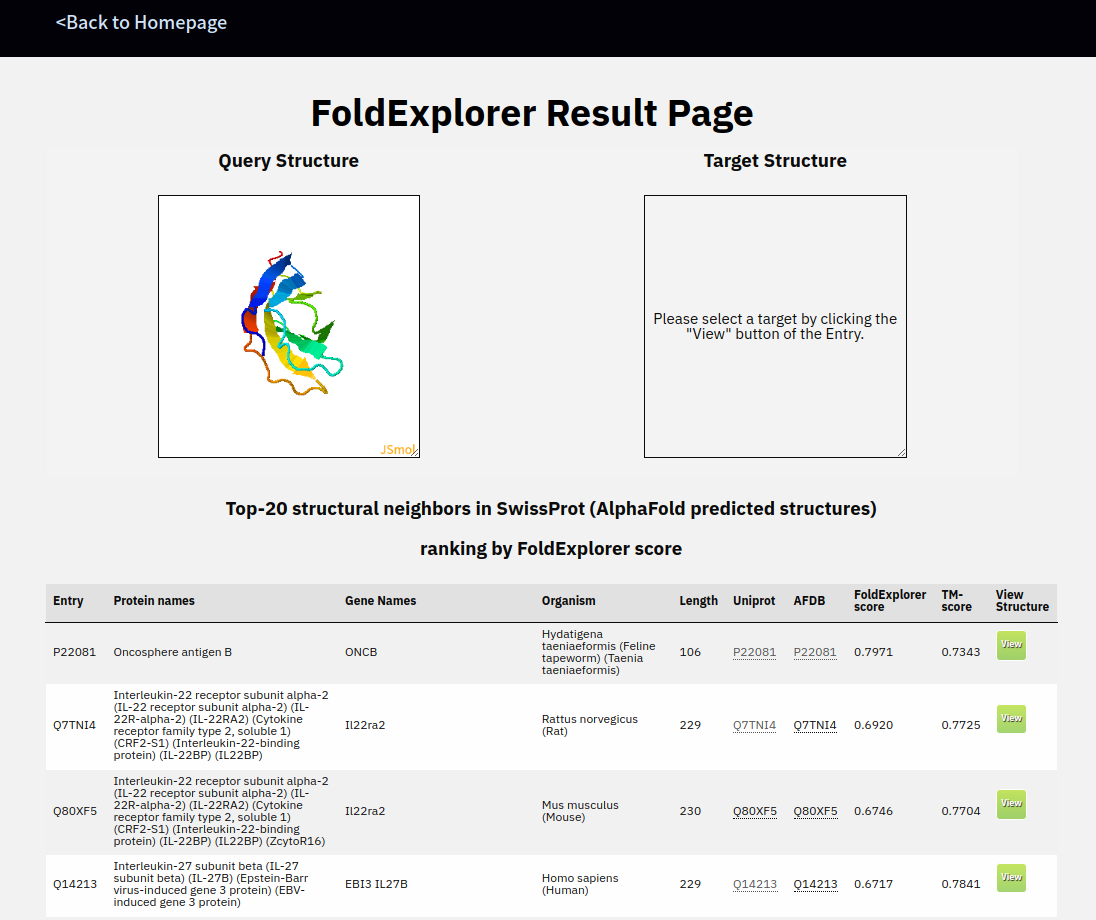
You can click the green view button and the 3D structure will be dispalyed in the "Target structure" canvas.
FoldExplorer harnesses the powerful capabilities of graph attention neural networks and protein large language models for protein structures and sequences data processing to generate embeddings for protein structures. The structural embeddings can be used for fast and accurate protein search. And the embeddings also provide insights into the protein space. FoldExplorer demonstrates a substantial performance improvement over the current state-of-the-art algorithm on the benchmark datasets. Meanwhile, FoldExplorer does not compromise on search speed and excels particularly in searching on a large-scale dataset.
The overall pipeline of FoldExplorer is shown as below.