|
RNAJog is a tool designed for optimizing the coding sequence (CDS) of mRNA to achieve high protein expression levels. RNAJog can generate codon sequences with high codon adaptation index (CAI) and low minimum free energy (MFE), ensuring enhanced translational efficiency and mRNA stability. This tool enables users to optimize mRNA sequences for their target proteins or existing mRNA sequences. The following guidance will assist you in generating your optimized mRNA sequences effectively. Note: * The software is free to academic users ONLY; For commercial usage, please contact with us. * Please do not use any crawler tools or submit frequently in a very short time . Guidance
Step 1: Select the Optimization Model
Step 2: Input the Sequence
Step 3: Configure Optimization Settings Output If protein is chosen as the input, the output file will contain the optimized codon sequences, along with their corresponding decoding strategy, sequence length, MFE-CAI weight, Minimum Free Energy (MFE), and Codon Adaptation Index (CAI). If RNA is chosen as the input, the output file will append the provided RNA sequence to the top of the table, with its decoding strategy labeled as "input" in the table. 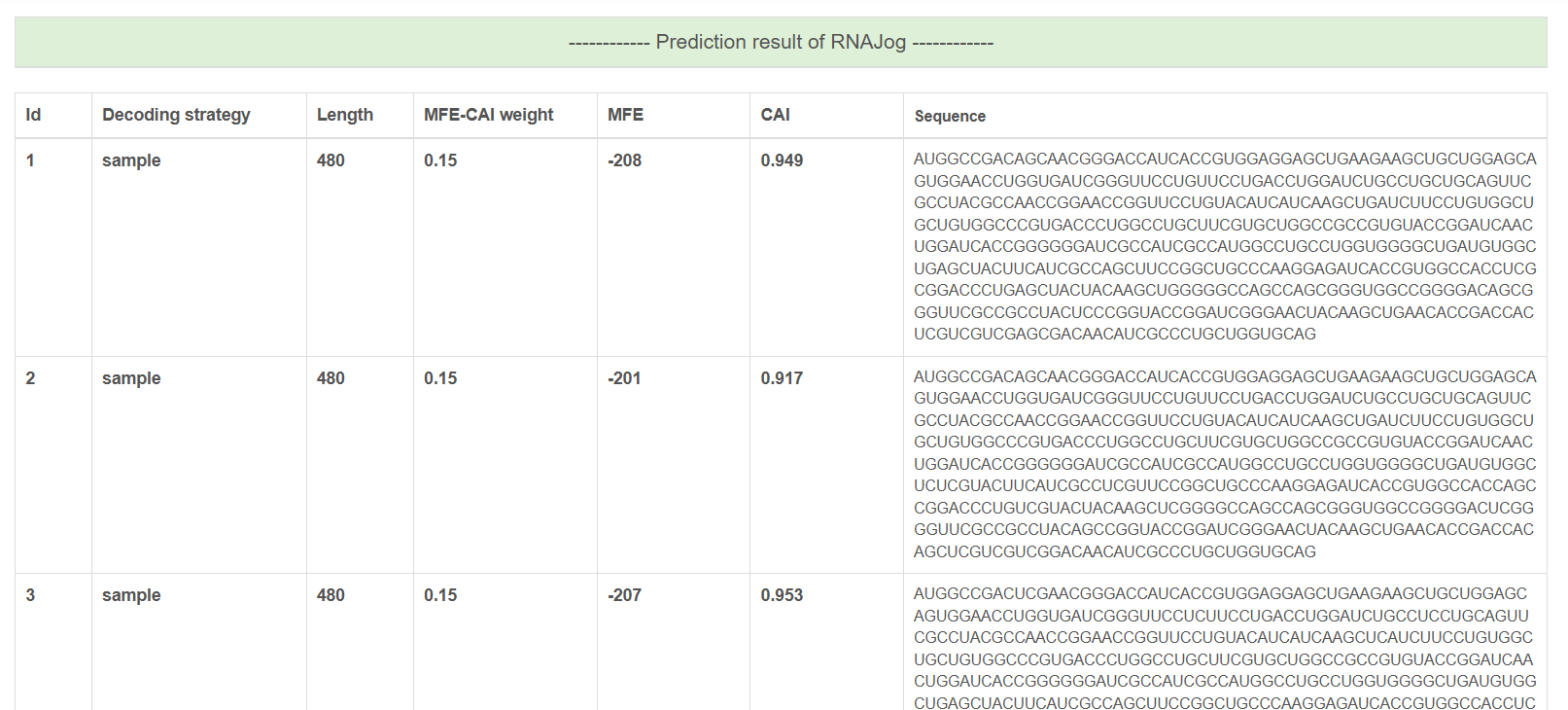
|