|
Incorporating label correlations into deep neural networks to classify protein subcellular location patterns in immunohistochemistry images
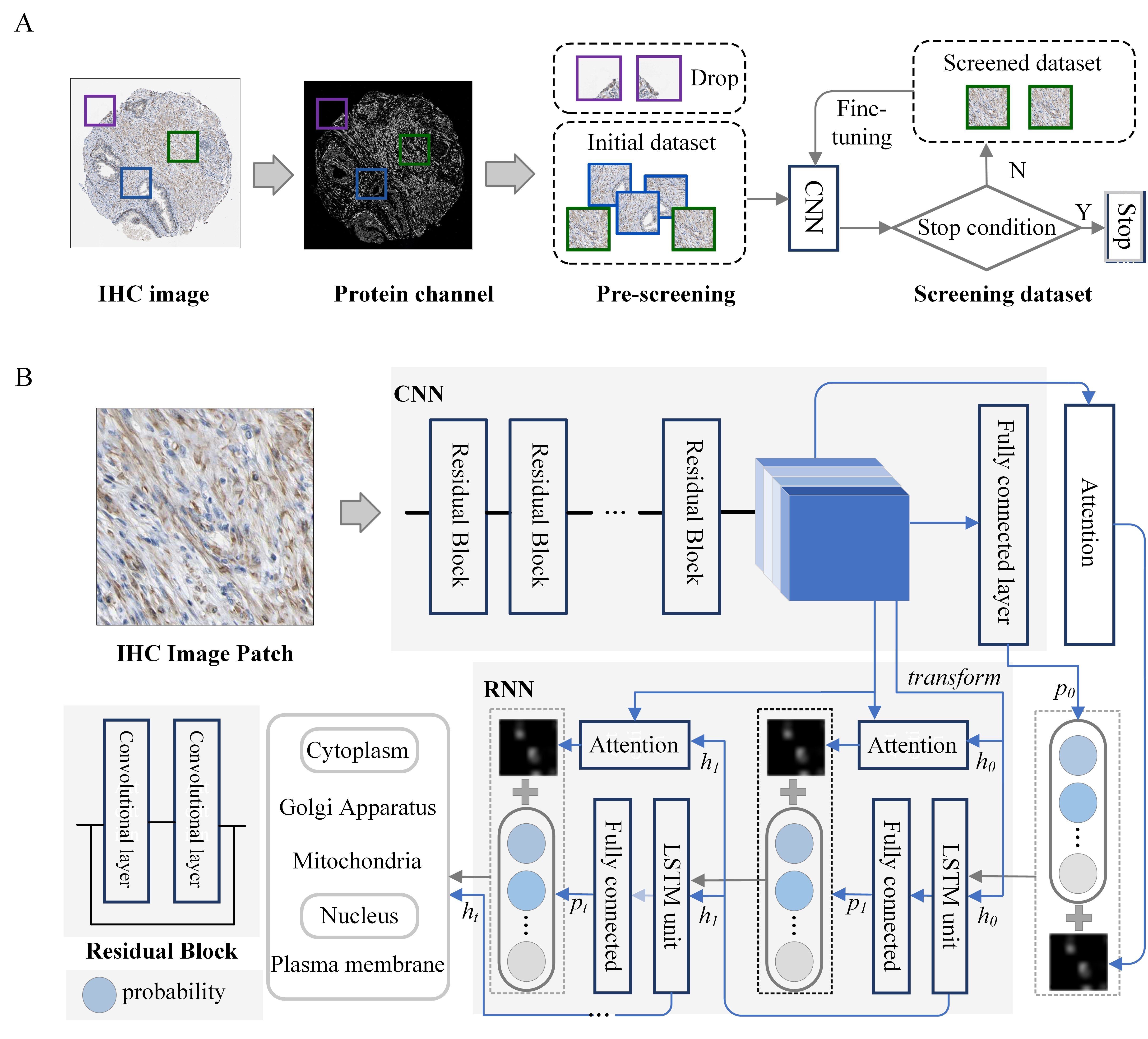
Introduction Analysis of protein subcellular localization is a critical part of proteomics. In recent years, as both the number and quality of microscopic images have increased rapidly, many automated methods, especially convolutional neural networks, have been developed to predict protein subcellular location(s) based on bioimages, but their performance always suffers from some inherent properties of the prediction problem. First, many microscopic images have non-informative or noisy sections, like unstained stroma and unspecific background, affecting the extraction of protein expression information. Second, the patterns of protein subcellular localization are very complex, as a lot of proteins localize in more than one compartment. In this study, we propose a new label-correlation enhanced deep neural network, laceDNN, to classify the subcellular locations of multi-label proteins from immunohistochemistry images. The model uses small representative patches as input to alleviate the image noise issue, and its backbone is a hybrid architecture of convolutional neural network and recurrent neural network, where the former network extracts representative image features and the latter learns the organelle dependency relationships. Our experimental results indicate that the proposed model can improve the performance of multi-label protein subcellular classification. Availability The laceDNN is available at www.csbio.sjtu.edu.cn/bioinf/laceDNN/.
Fig. 1. Flow chart of laceDNN. Reference Jin-Xian Hu, Yang Yang, Ying-Ying Xu, Hong-Bin Shen, Incorporating label correlations into deep neural networks to classify protein subcellular location patterns in immunohistochemistry images, Submitted. |