|
Readme |
|
|
|
DELIA is a web server designed for structure-based protein-ligand binding site prediction. Multiple deep neural networks are combined to generate the binding probability for each residue in a single protein chain. Distance matrices extracted from 3D coordinates are integrated with sequence-profiling as the input features of DELIA. DELIA has been evaluated on several benchmark datasets and the results show that it can outperform its old version and state-of-the-art methods. |
| Result Page |
|
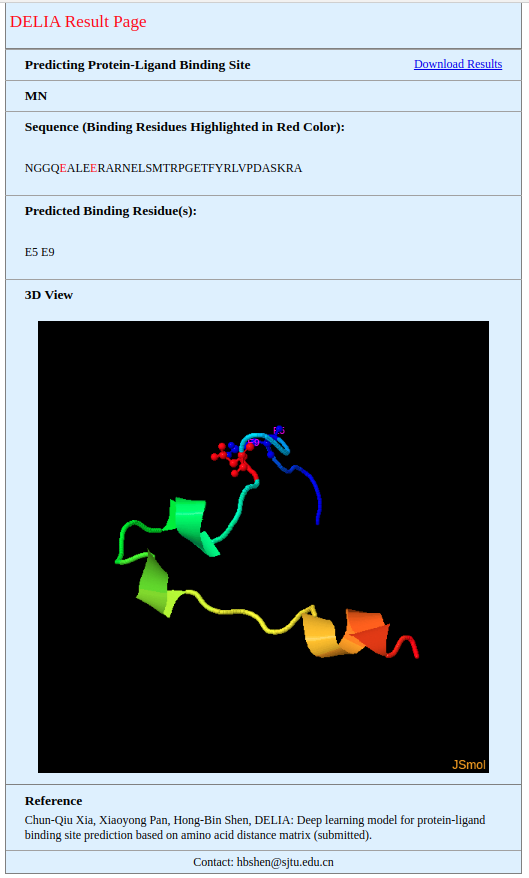
Results will be shown in the result page when the job is finished. The example result page is shown in the figure below. Binding residues are marked as red in the sequence extracted from the PDB file. The 3D structure of query protein and binding residues are also shown in the JSmol. In addition, results can be downloaded by clicking the 'Download Results'. |
|
|
| Q&A |
|
1. How should I use DELIA?
First, you need to upload a PDB format file or paste the text which contains 3D coordinates of your query protein; second, you need to designate the target chain ID because our model can only be applied to single chain; third, select the ligand type; at last, you can input the email address which is used to receive the results. |
|
2. How long will it take to generate the results?
In fact, running time is related to the length of query protein chain. About additional 100 residues in the query sequence may increase the running time by one hour. Therefore, We do not recommend you to submit a very long protien chain. |
|
3. How many proteins can I submit per day?
We recommend you to submit no more than 10 PDB file per day due to the limitation of our computing resources. However, if your query sequences are relatively short (e.g., shorter than 200 residues), you can submit a little more. |