The following points are important when using Signal-CF
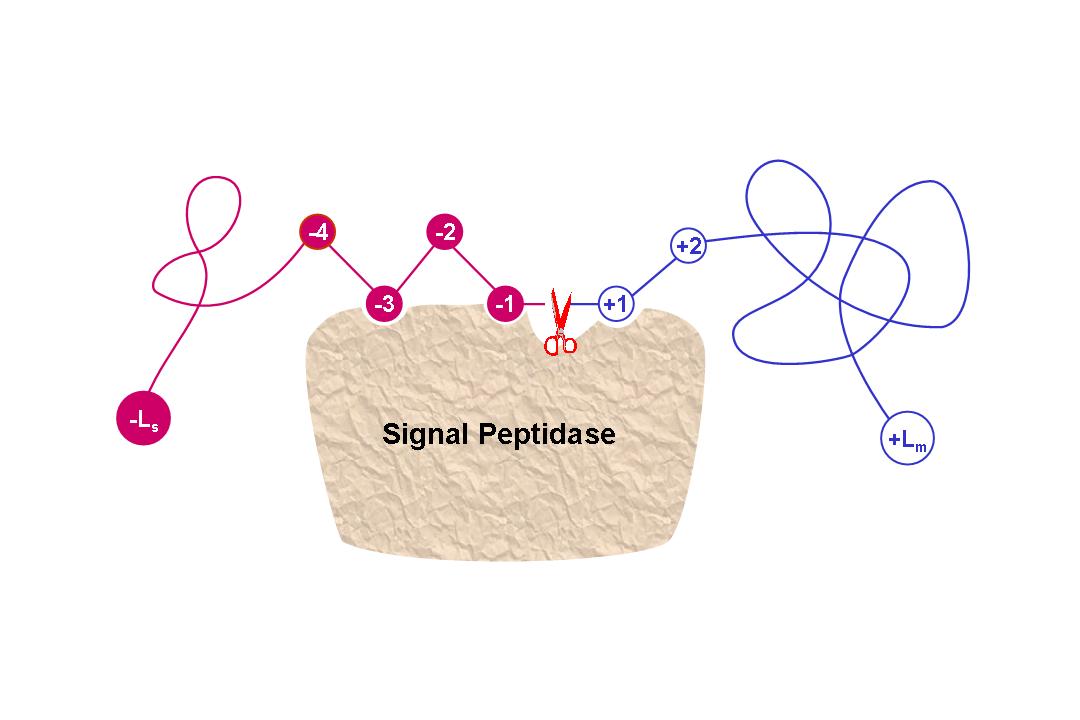
1. Signal-CF is focused on the prediction of N-terminal signal peptides only (see the figure below).
2. Your input sequence length should be at least more than 50aa counted from the N-terminal.
3. Signal-CF is designed for "eukaryotic","Gram-positive" and "Gram-negative" proteins. Please select the correct organism type before you start computation.