| Introduction of Signal-3L 2.0 |
|
Signal-3L 2.0 is an online server for predicting the N-terminal protein signal peptide, and the input is the amino acid sequence only. It is constructed with a hierarchical mixture model, which contains the following three layers (Figure 1): (1) Discrimination of SP (Signal Peptide) proteins and TMH (TransMembrane Helical) proteins from the other globular proteins; (2) Recognizing SP proteins from TMH proteins; (3) Identifying the cleavage sites of SP proteins. |
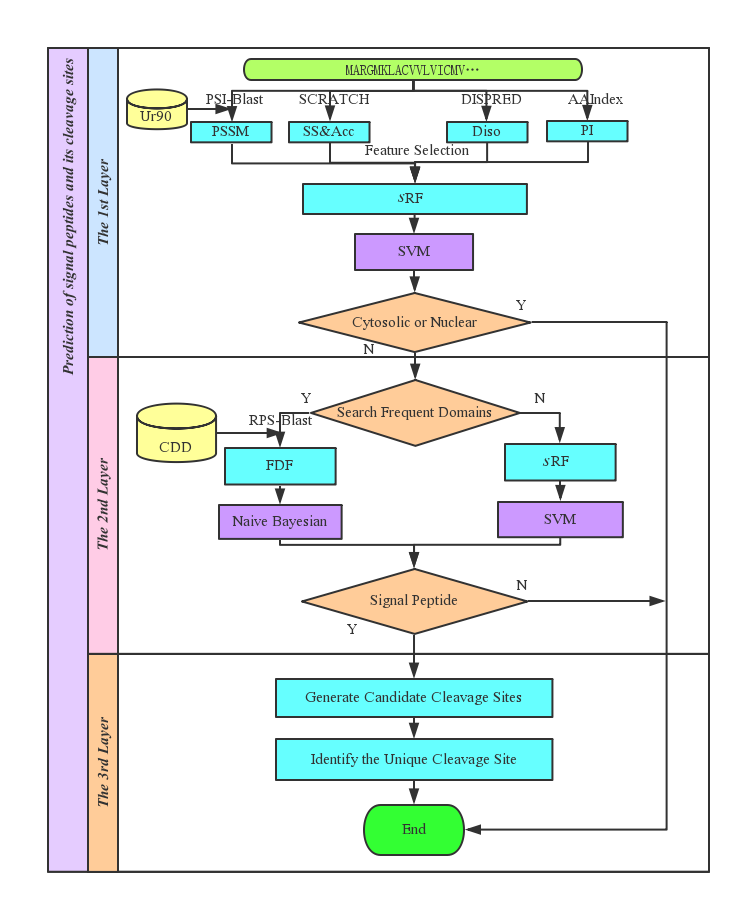 |
| Figure 1. The flowchart of Signal-3L 2.0. |
Examples of the output of Signal-3L 2.0 are shown in Figure 2,3,4, which will give the scores of several candidate cleavage sites: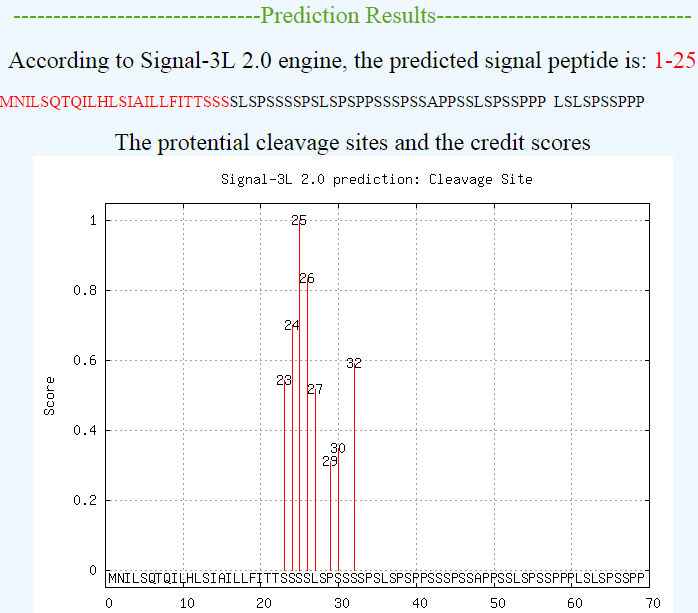
|
| Figure 2. An example to show the output of Signal-3L 2.0 for a signal peptide protein. |
 |
| Figure 3. An example to show the output of Signal-3L 2.0 for a protein Not of a signal peptide or TMH membrane protein. |
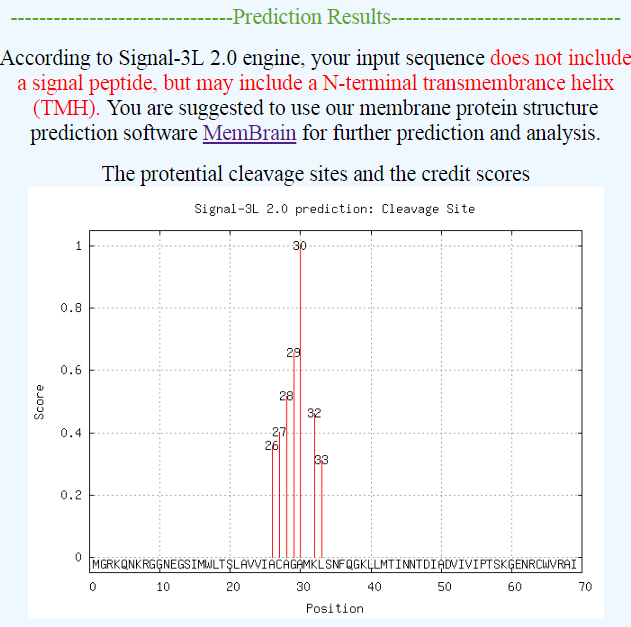 |
| Figure 4. An example to show the output of Signal-3L 2.0 for a TMH membrane protein. |
|
Note: The following points are important when using Signal-3L 2.0: 1. The servier is focused on the prediction of N-terminal signal peptides only. 2. It is designed for "Eukaryotic ","Gram-positive" and "Gram-negative" proteins. Please select the correct organism type before you start computation. |