|
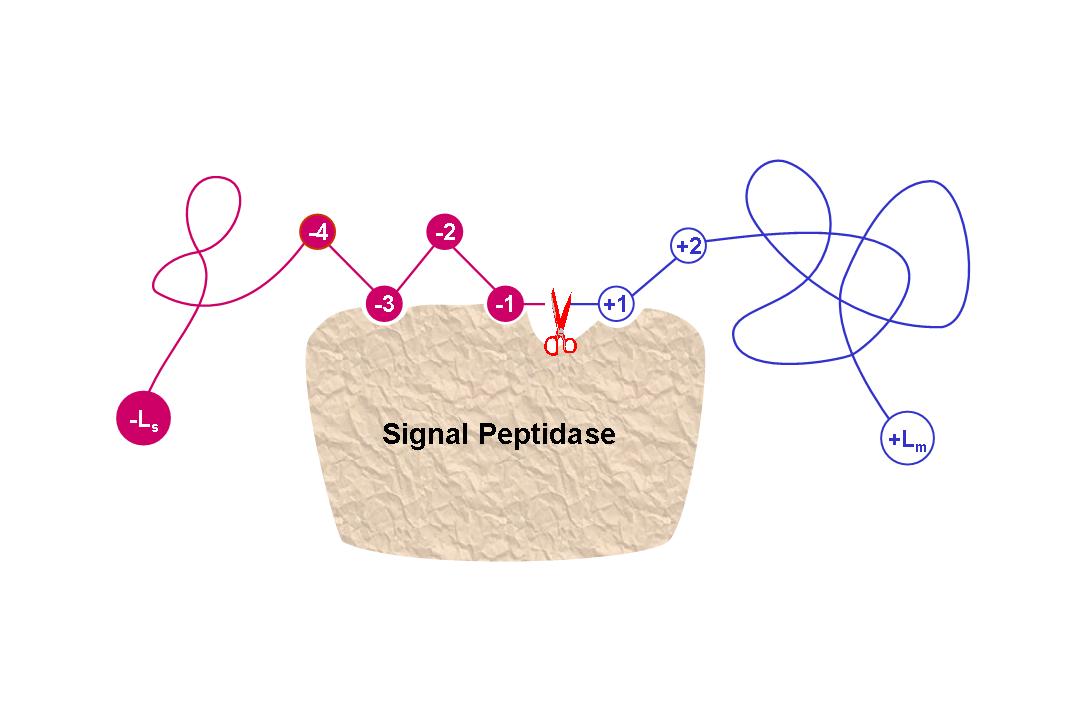
Signal-3L is an automated method for predicting
signal peptide sequences and their cleavage sites in eukaryotic and bacterial protein sequences. It consists of three prediction engines working respectively for the following three
progressively deepening layers: (1) Identifying a query protein as secretory or non-secretory by an ensemble classifier; (2) Selecting a set of candidates for the possible signal peptide cleavage sites of a query secretory protein by a subsite-coupled discrimination algorithm; (3) Determining the final cleavage site by fusing the global sequence alignment outcome for each of the aforementioned candidates through a voting system. |
| Caveat |
|
The following points are important when using Signal-3L 1. Signal-3L is focused on the prediction of N-terminal signal peptides only (see the figure below). 2. Your input sequence length should be at least more than 50aa counted from the N-terminal (see the figure below). 3. Signal-3L is designed for "human","plant","animal","other eukaryotic","Gram-positive" and "Gram-negative" proteins. Please select the correct organism type before you start computation. |
 |