|
Bioimage-based Semi-supervised Method for Protein Subcellular Localization
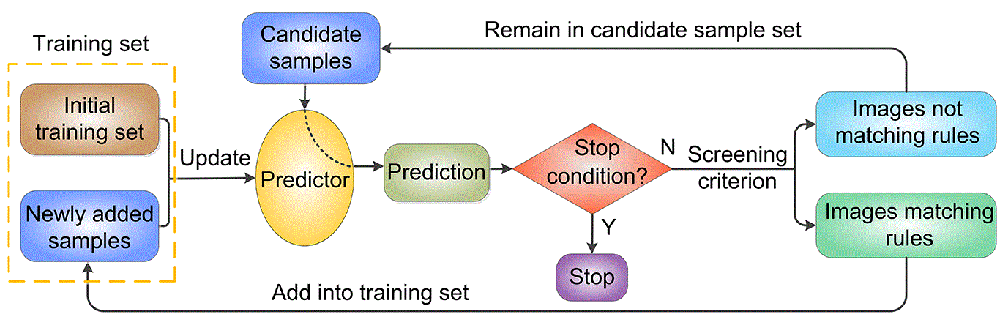
Introduction There is a long term interest in the challenging task of finding translocated and mislocated cancer biomarker proteins. Bi-oimages of subcellular protein distribution are new data sources for this task which have attracted much attention in recent years because of their intuitive and detailed descriptions of protein distribution. However, automated methods in large-scale biomarker screening suffer significantly from the lack of subcellular location annotations for bioimages from cancer tissues. The transfer prediction idea of applying models trained on normal tissue proteins to predict the subcellular locations of cancerous ones is arbitrary because the protein distribution patterns may differ greatly in normal and cancerous states. We developed for the first time a new semi-supervised protocol that can use unlabeled cancer protein data in model construction by an iterative and incremental training strategy. Our approach enables us to selectively use the low-quality images in normal states to expand the training sample space and provides a general way for dealing with the small size of annotated images used together with large unannotated ones. Experiments demonstrate that the new semi-supervised protocol can result in improved accuracy and sensitivity of subcellular location difference detection. A flow chart of the semi-supervised method is shown in Figure 1.
Fig. 1. Flow chart of the iterative incremental training process. Code and dataset The data and code are contained in the following compressed files: Click here to download the datasets (1.03Gb), and click here to download the source code (229Kb). The code package has been tested using Matlab 2011b under Windows 7 in a 64bit architecture. Reference Ying-Ying Xu, Fan Yang, Yang Zhang, Hong-Bin Shen, Bioimaging based detection of mislocalized proteins in human cancers by semi-supervised learning, Bioinformatics, 2015, 31: 1111-1119. |