
SRes:Resolution measurement from a single reconstructed Cryo-EM density map with multi-scale spectral analysis
Introduction
As a relatively new protocol to solve the 3D structure of a protein or protein complex, Single-Particle Reconstruction (SPR) of cryo-EM images shows much superiority and is in a rapidly developing stage. Resolution measurement in SPR, which evaluates the quality of a reconstructed 3D density map, plays a critical role for promoting methodology development of SPR and structural biology. Due to there is no benchmark map in a new structure generation, how to realize the resolution estimation of a new map is still an open problem. Existing approaches try to generate a hypothetical benchmark map by reconstructing two 3D models from two halves of the original 2D images for cross-reference, which may result in a premature estimation with a half-data model.
In this paper, we report a new self-reference-based resolution estimation protocol (SRes), which only requires a single reconstructed 3D map. The core idea in SRes is performing a multi-scale spectral analysis (MSSA) on the map through multiple size-variable masks segmenting the map. The MSSA-derived multi-scale spectral signal-to-noise ratios (mSSNR) reveal their corresponding estimated resolutions will show a cliff jump, indicating a significant change of the SSNR properties. The critical point on the cliff border line is demonstrated as the right estimator for the resolution of the map.
The flow chart of SRes is shown in Figure 1.
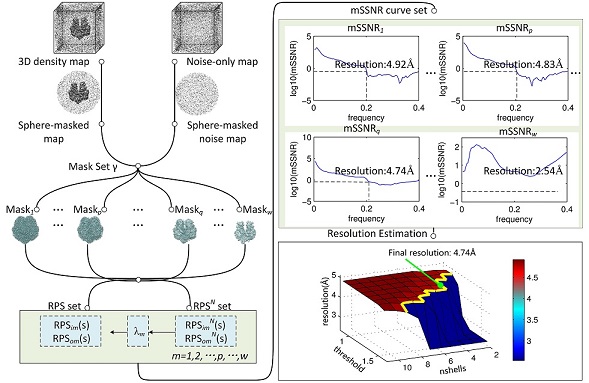
Fig 1. Flowchart of SRes
Source code
The source code and example data are contained in the following compressed files:SRes.tar.gz
The code package has been tested using Matlab 2014a under ubuntu 14.04 LTS in a 64bit architecture.
Reference
Yu-Jiao Yang, Shuai Wang, Biao Zhang, and Hong-Bin Shen, resolution measurement from a single reconstructed Cryo-EM density map with multi-scale spectral analysis, Journal of Chemical Information and Modeling, 2018 (in press).