ATTENTION : The software is free to academic users ONLY; For commercial usage, please contact with us.
Please do not use any crawler tools or submit frequently in a very short time. (especially general model)
Due to the limitation of computing resources, we temporarily support prediction for files below 500Kb.
Manual of RBPsuite webserver
RBPsuite is an deep learning based approach suite to predict RNA-binding protein (RBP) sites on linear and circular RNAs
Content
Output
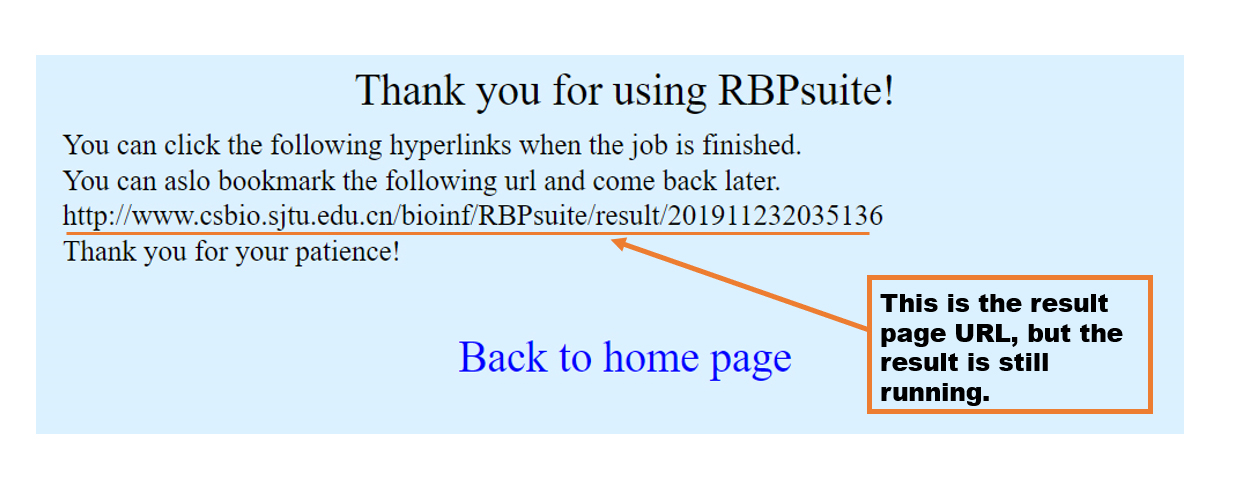
It will take a while for program to run. The result page URL is given but the result hasn't been finished, so you can bookmark this URL and come back later or wait for a while.
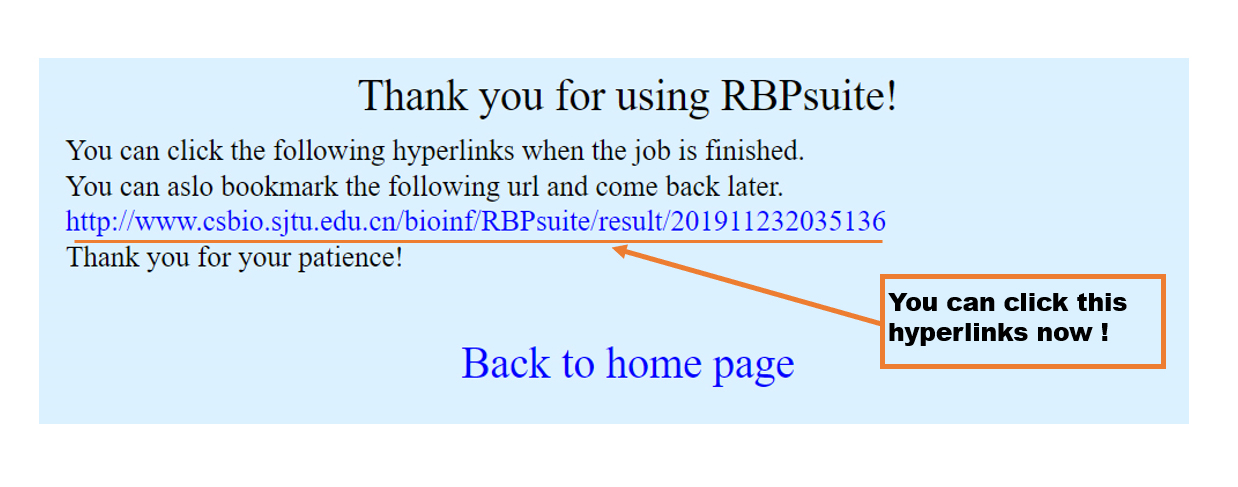
After a while(about 10 seconds), the result page is ready and you can click this URL to the result page.
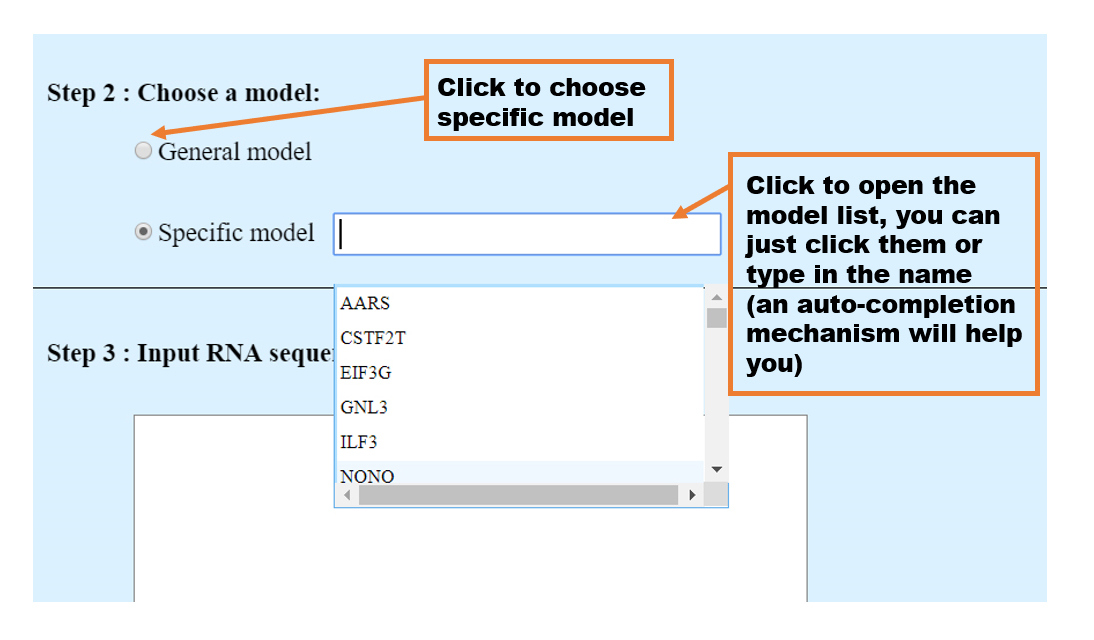
1. Specific model
If the RNA-binding protein has a verified motif, RBPsuite will provide the motif logo, which will be visualized along the input sequence.

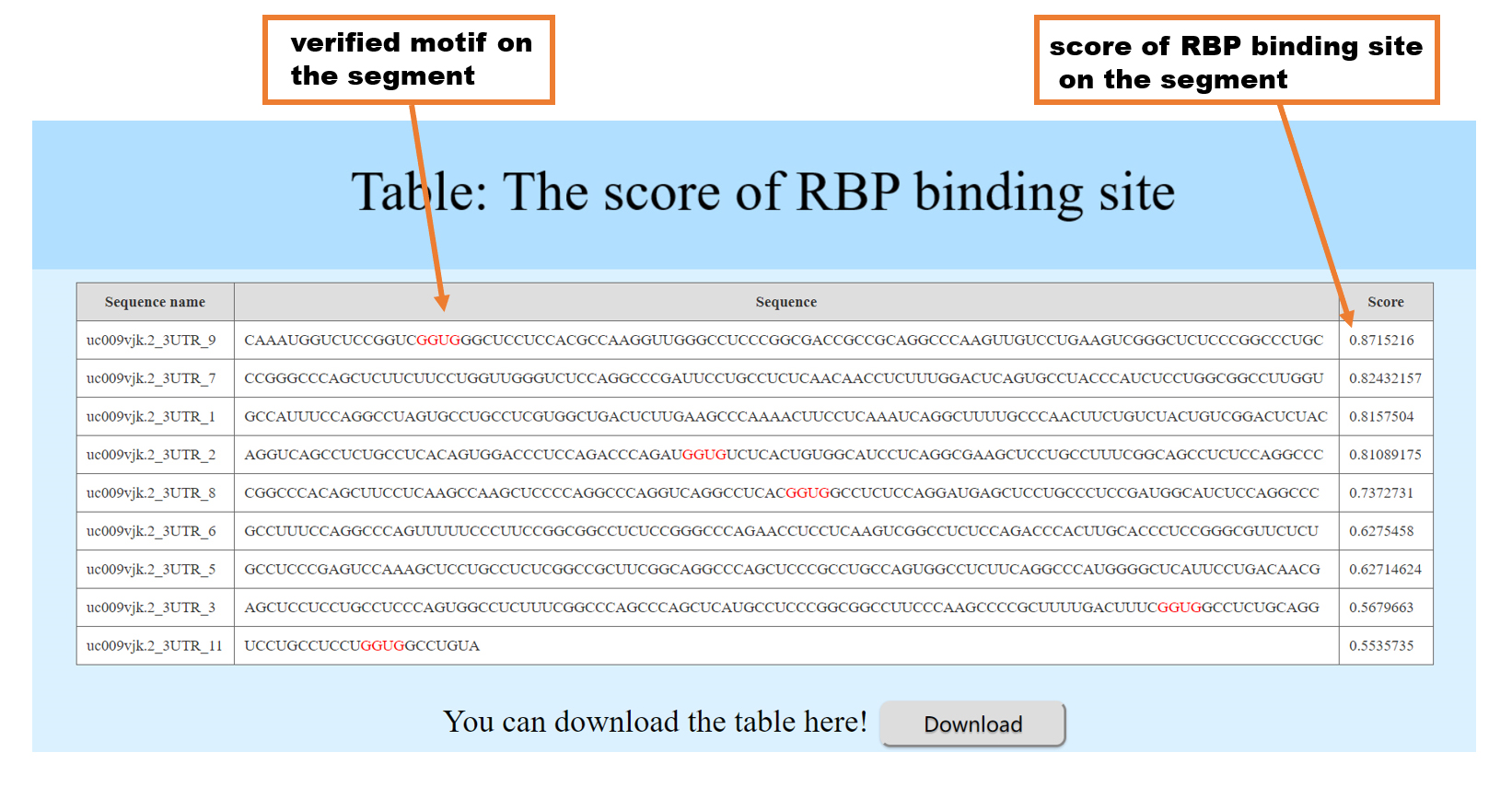
In order to improve the efficiency of the calculation , RBPsuite divides the input sequence into segments of length 101 without overlap and display the score of each segment with the RBP of interest in the table. In addition, RBPsuite has numbered the sequence segment according the orders in the input sequence like _1,_2,_3... in the first column of the table, _1 means the first 101nt segment, _2 means the second 101nt segment and so on, along the input sequence. These segments are arranged in descending order of score, and segments with a score less than 0.5 are filtered out. When the RBP has a verified motif file, for the segments with a score greater than 0.5, RBPsuite marks the possible motifs in RED according to the position on the segment. (using the MEME-fimo tool: http://meme-suite.org) What's more, RBPsuite provides sort function. You can click the name of each column to sort the results ascendly and descendly. (the "sequence" column is not supported)
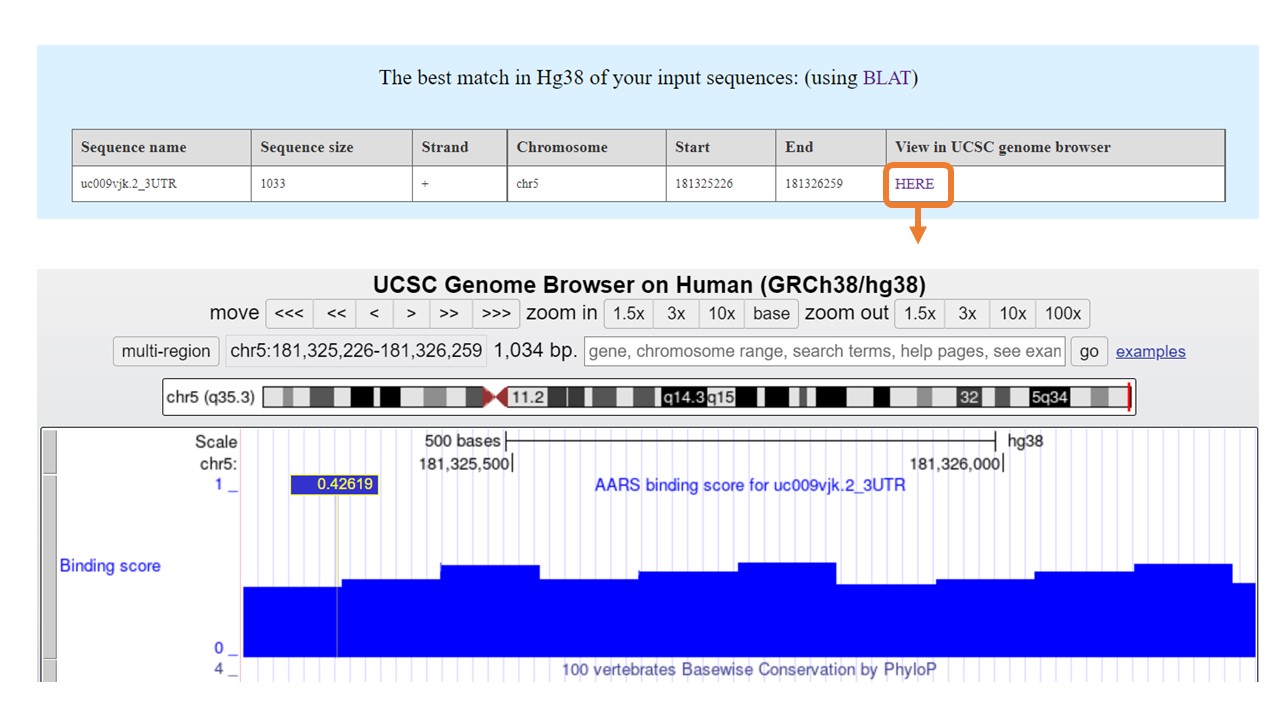
RBPsuite provides the best match of the input sequence in Genome Reference Consortium Human Build 38(hg38) by BLAT. The detailed information includes sequence length, chromosome, strand, start index, and end index. Besides, RBPsuite visualizes the input sequence with the binding score track(prediction score on the sequence) by the UCSC genome browser.
RBPsuite also provides visualization for the binding scores of the segments along the input sequence, it displays the scores in a direct way.

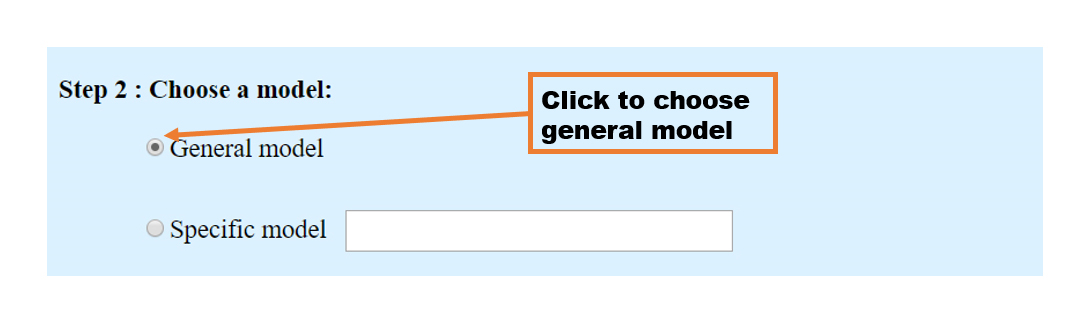
2. General model
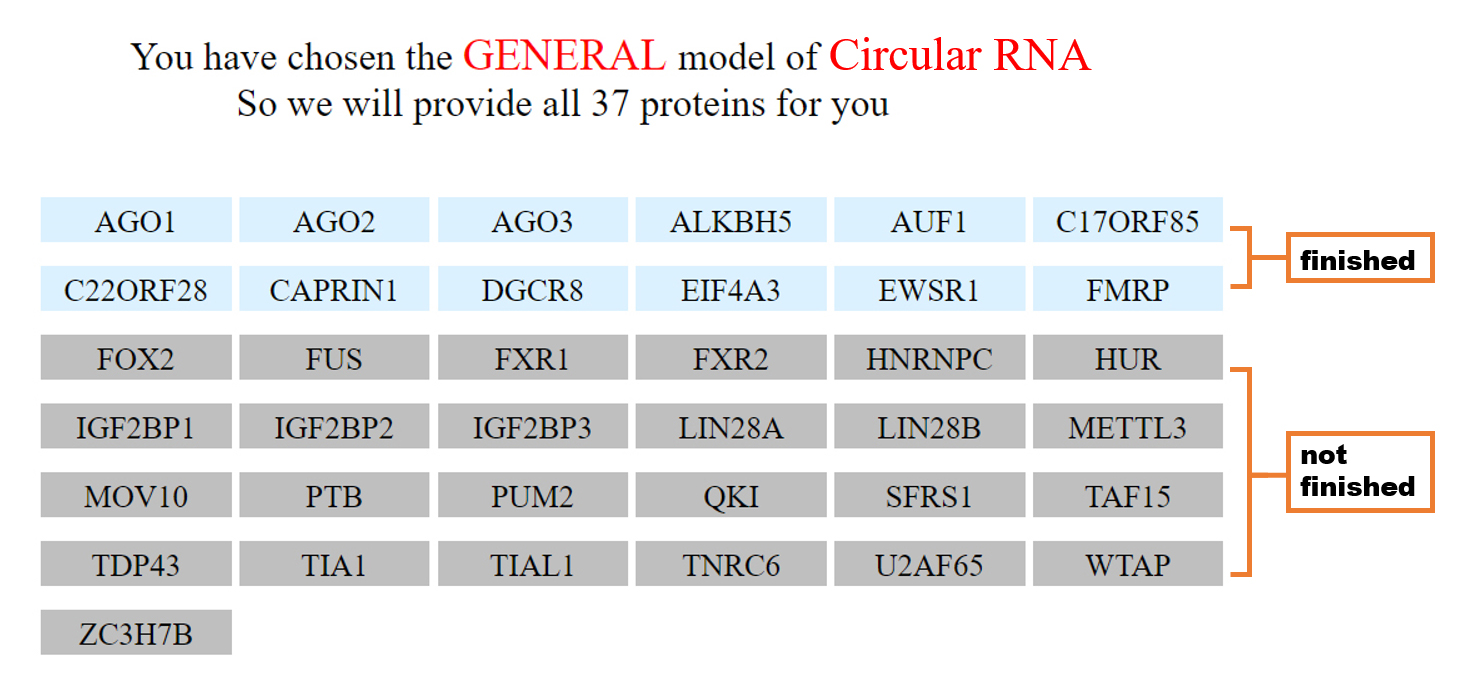
Different from the specific model, the general model has a directory interface that lists all the RBPs, which predict binding scores between your input RNA and the model trained for this RBP. The results that have been finished are displayed in blue, and those displayed in gray means not finished yet. For more details about the prediction results of individual RBPs, the users can click the RBP of interest to see the predicted RBP binding sites of this RBP for the input sequence. The result page of each RBP is the same as the specific model.
Q&A
Question: The result page URL stays grey and accessing it results in a 403 Forbidden error.
Answer: Please check if your input file is in FASTA format.
Question: The result page does not show any table or figure.
Answer: Recently some resources on our website server are unavailable to access by off-campus IP addresses.If you are unable to view the results, you can send an email to the temporary email address 'fy826943194@sjtu.edu.cn', and attach the URL of the result like 'http://www.csbio.sjtu.edu.cn/bioinf/RBPsuite/result/202305270004130'. We will send the results back to your email.