|
Hum-mPLoc 3.0 Tutorial
|
| 1. Introduction | ||||||||
|
Protein subcellular localization has been an important research topic in computational biology
over the last two decades. Till now, a variety of computational methods have been proposed to deal
with large scale data sets of proteins with unknown locations. The statistical machine learning-based
approaches are a major branch of the existing predictors, and tremendous studies have shown that
features extracted from biological domain knowledge can be very useful for improving the prediction
accuracy. However, the domain knowledge, such as Gene Ontology and functional domain, usually results
in redundant features and high-dimensional feature spaces, which may degenerate the performance of
machine learning models. We propose a new feature representation protocol denoted as HCM (Hidden Correlation Modeling). The HCM method is featured by considering structural hierarchy of the domain knowledge base and the correlations between annotation terms, so as to create more compact and discriminative feature vector. Hum-mPLoc 3.0 is a web server developed for human protein subcelluar prediction. The predictor covers the following 12 subcellular locations: (1) centriole, (2) cytoplasm, (3) cytoskeleton, (4) endoplasmic reticulum, (5) endosome , (6) extracell, (7) Golgi apparatus, (8) lysosome, (9) mitochondrion, (10) nucleus, (11) peroxisome, and (12) plasma membrane. | ||||||||
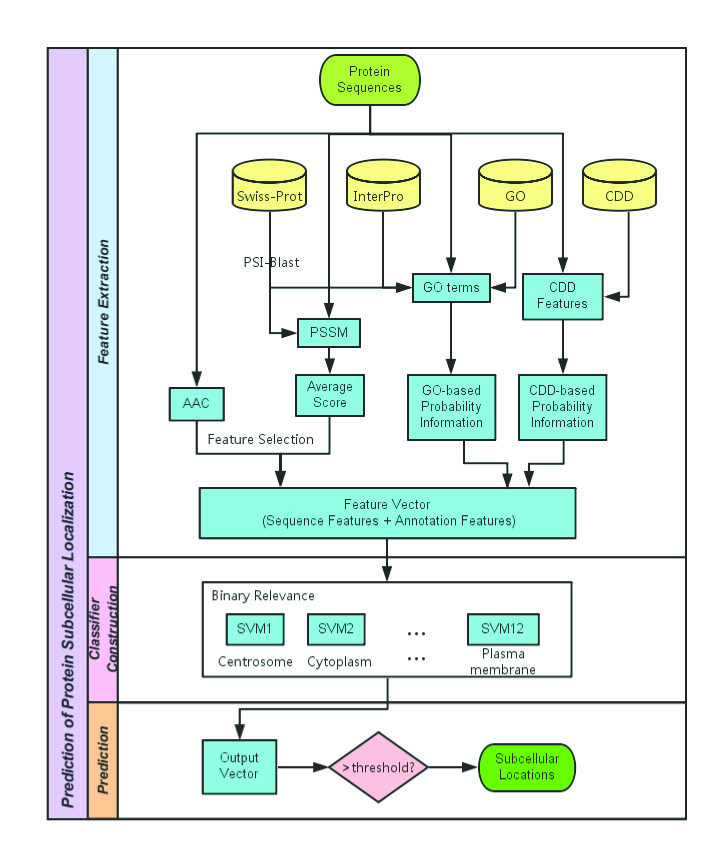 Figure 1. Flowchart of subcellular localization prediction in Hum-mPLoc 3.0. |
||||||||
|
|
| 2. Inputs | |
|
(A). Protein sequence Input the protein sequence into the input box in Fasta format. | |
|
(B). GO sources If you select "Experimental", Hum-mPLoc 3.0 will only select GO annotations with experimental evidence for prediction. If you select "All sources", Hum-mPLoc 3.0 will select all GO annotations assigned by Gene Ontology curators for prediction | |
|
(C). Email address You should input your email address to receive an email notification of your prediction results. | |
| 3. Outputs | |
|
The prediction result and each subcellular localization's decision value will be sent to your email. For the convenience of users to get a quick search, we pre-computed results for all human proteins in Swiss-Prot (release in May 2015), which contains total 20197 human proteins. The results can be downloaded by clicking Whole Human Proteome Prediction. You can also submit the protein's AC (Accession Number) or ID (Name) to retrieve the results for a specific protein target in page Whole Human Prediction. According to Hum-mPLoc 3.0 model, 16717 proteins are predicted locating in one location, 3104 proteins locating in two locations, 335 proteins locating in three locations and 41 proteins locating in four locations. | |