|
Readme |
|
|
|
GraSR is a web server designed for protein structure representation and fast protein structure retrieval. Deep Graph neural networks are designed to represent the 3D coordinate of protein structures. It was trained on SCOPe v2.07 (40% ID filtered subset) with a momentum contrastive learning framework. GraSR has been cross-validated on the aforementioned SCOPe dataset and tested independently on a subset of PDB released recently. The results show that it outperforms the state-of-the-art methods. Our server follows a "filter and refine" paradigm to improve the retrieval accuracy. At first, GraSR is used to filter similar structures from the database fastly. Then, TM-align is used to derive the TM-score of these selected structures. Finally, we re-rank them according to the TM-score and show the Top-20 similar structures. |
| Result Page |
|
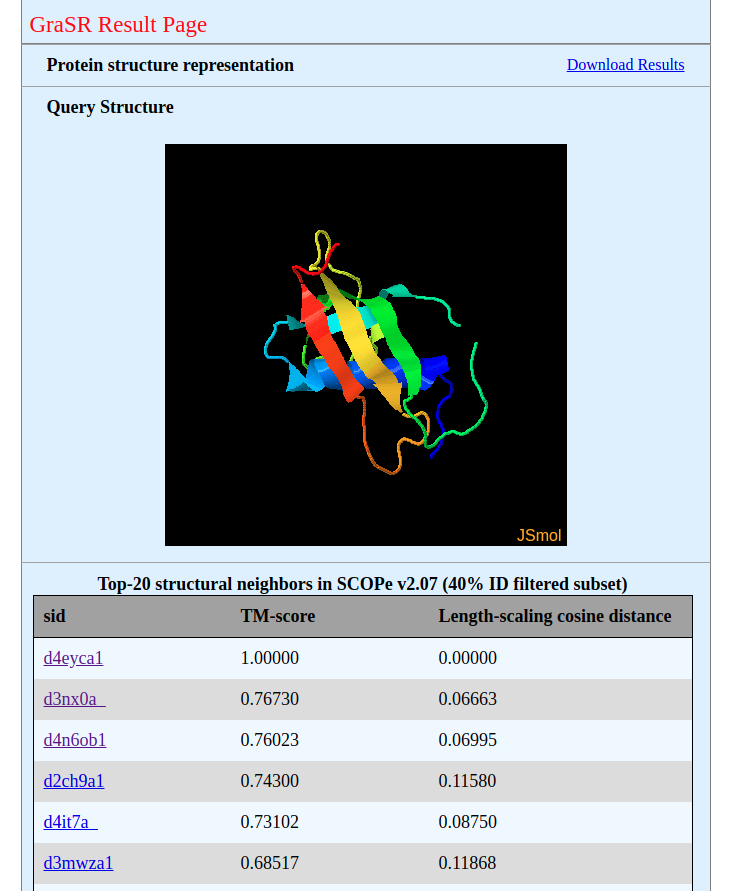
Results will be shown in the result page when the job is finished. The example result page is shown in the figure below. If you choose a database, twenty most similar structures in it to the query structure will be shown; otherwise, only the descriptor of the query structure will be generated and can be downloaded by clicking the 'Download Results'. |
|
|
| Q&A |
|
1. How should I use GraSR?
First, you need to upload a PDB format file or paste the text which contains 3D coordinates of your query protein; second, you need to designate the target chain ID because our model can only be applied to single chain; third, select the database; at last, you can input the email address which is used to receive the results. |
|
2. How long will it take to generate the results?
Running time is related to the number of structures in the database. For SCOPe v2.07 (40% ID filtered subset), It will take about one minute. If you only need the descriptor of the query structure, it may take about 20 seconds. |