|
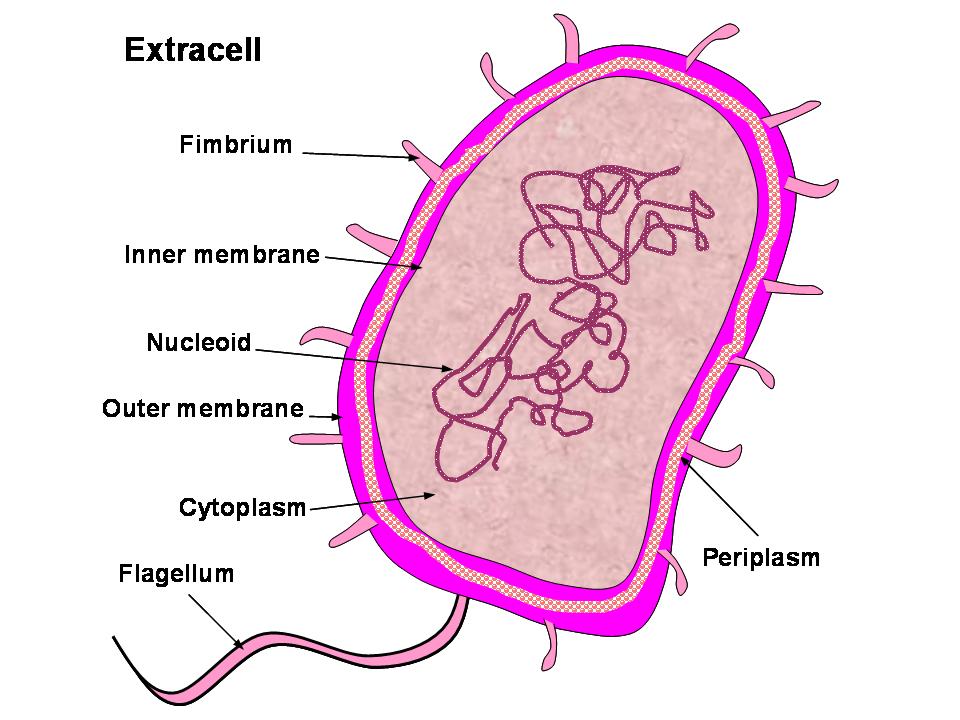
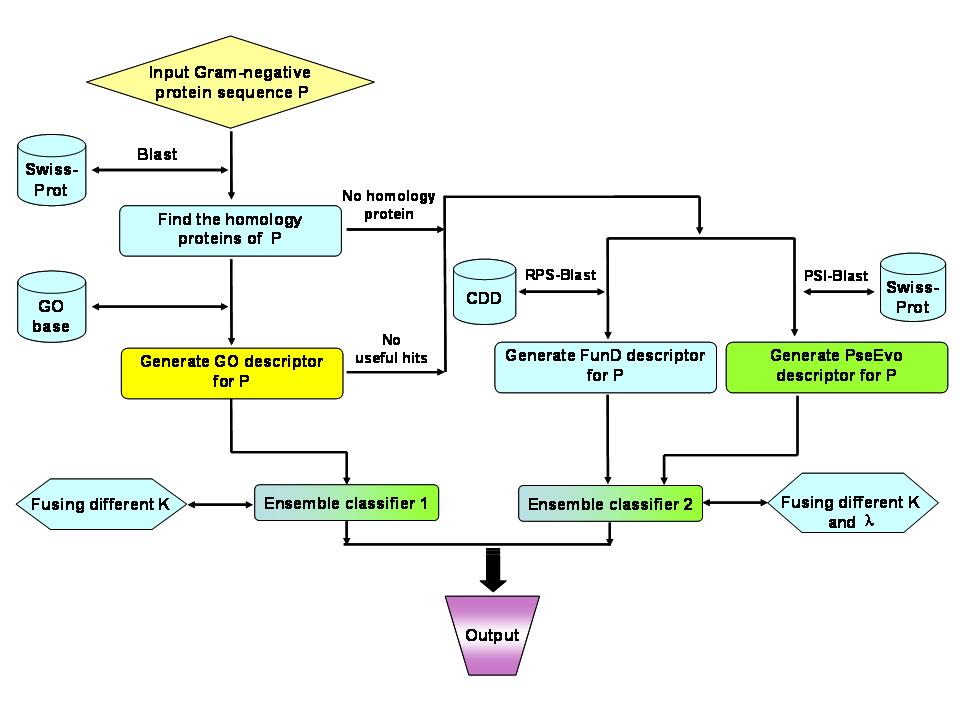
Gneg-mPLoc was developed for identifying the subcellular localization of Gram negative bacterial proteins by fusing the information of gene ontology, as well as the functional domain information and sequential evolution information. Compared with the old Gneg-PLoc, the new predictor is much more powerful and flexible. Particularly, it also has the capacity to deal with multiple-location proteins as indicated by the character "m" in front of "PLoc" of its name. The current version of Gneg-mPLoc can be used to identify the subcellular location of a query protein among the following eight sites: (1) cytoplasm, (2) extracell, (3) fimbrium, (4) flagellum, (5) inner membrane, (6) nucleoid, (7) outer membrane, and (8) periplasm (Figure 1). For the procedures of how Gneg-mPLoc works, see Figure 2.
|
| |
 |
| |
Figure 1. Schematic illustration to show the eight subcellular locations of Gram-negative bacterial proteins: (1) cytoplasm, (2) extracell, (3) fimbrium, (4) flagellum, (5) inner membrane, (6) nucleoid, (7) outer membrane, and (8) periplasm. |
| |
 |
| |
Figure 2. Flowchart to show how Gneg-mPLoc works. |

