Gaussian kernel is a popular kernel function often used in various computational biology and their applications to measure the similarity
between two samples in a dataset. It can be used not only in the unsupervised situation but also in the supervised case. However, the scalar parameter belta (¦Â) in Gaussian kernel function as shown in the following equation significantly affects the final results. This
online data-driven tool is used to calculate the optimal belta (¦Â) parameter based on the given dataset.
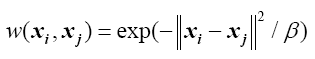
Note:
(1) When using this online service, the user is required to submit their dataset with a file named *.txt, for example "in.txt" .
(2) For very large-size or very high-dimensional datasets, the users need to download the stand alone program. |
| |
|
| |
|
|
| |
| |
| References: |
Jian-Bo Lei, Jiang-Bo Yin, and Hong-Bin Shen, GFO: A data driven approach for optimizing Gaussian function based similarity metric in computational biology, Neurocomputing, 2013, 99: 307-315. |
Jiang-Bo Yin, Tao Li, and Hong-Bin Shen, Gaussian kernel optimization: complex problem and a simple solution, Neurocomputing, 2011, 74: 3816-3822.
|
| |
|
Contact @ Hong-Bin |
|