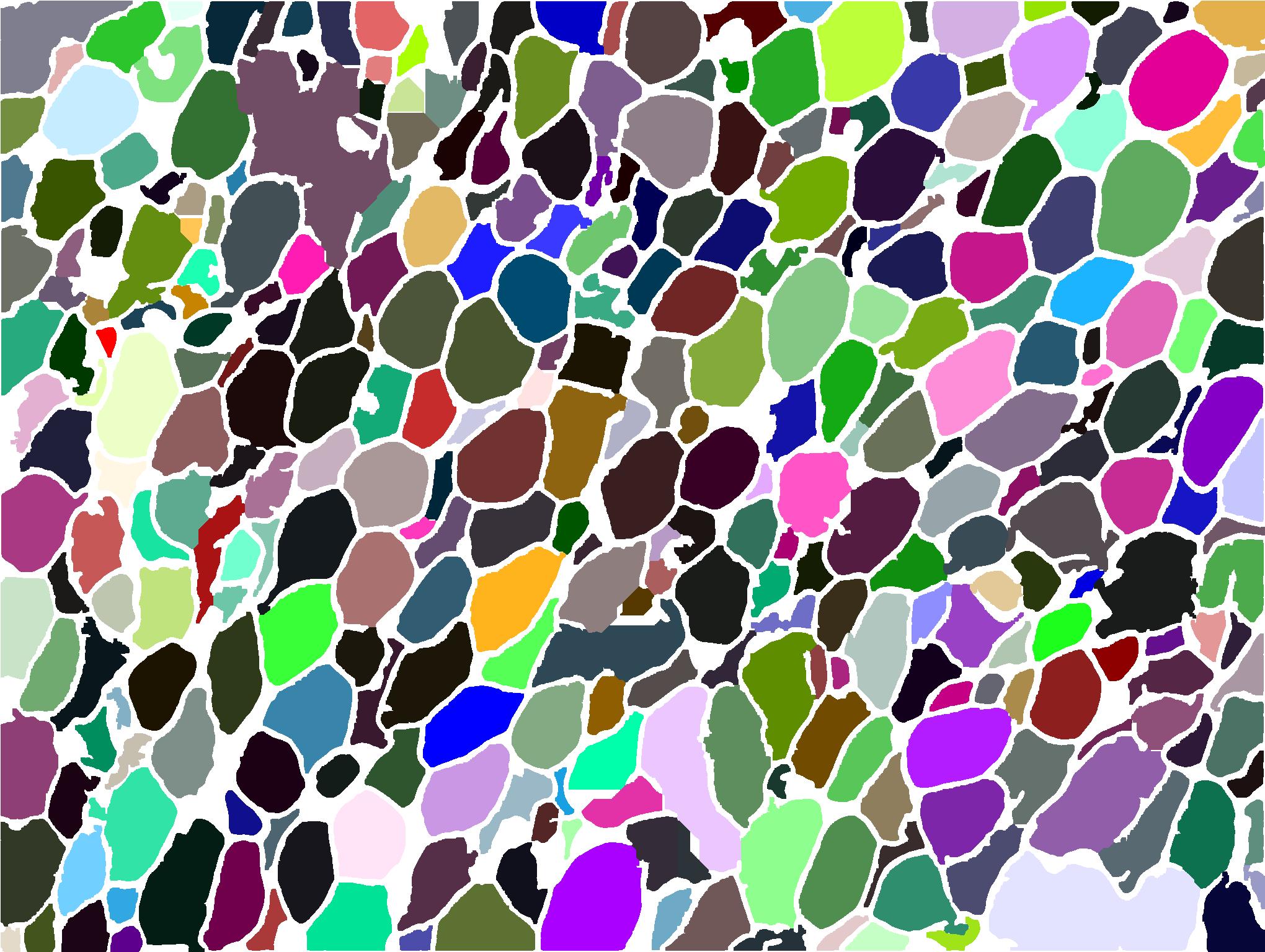
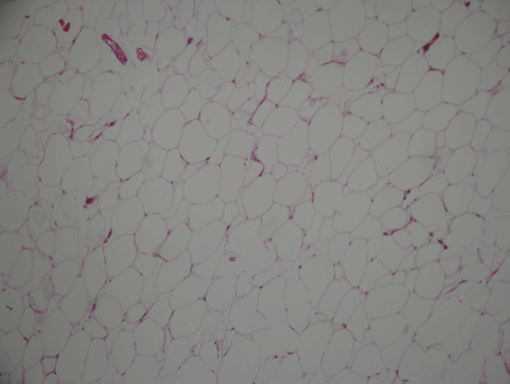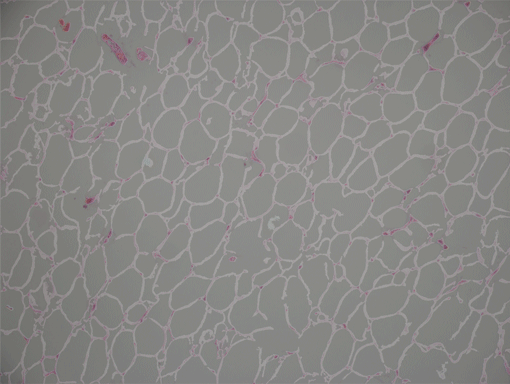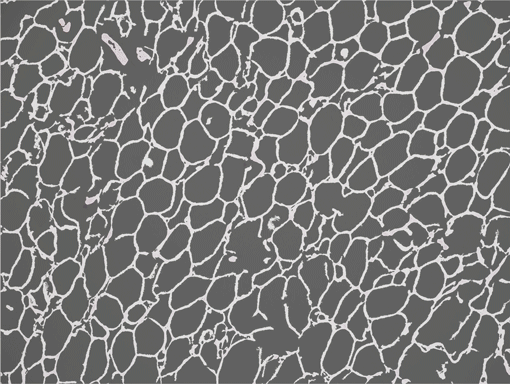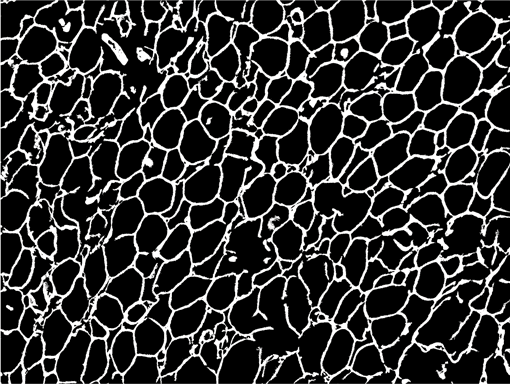
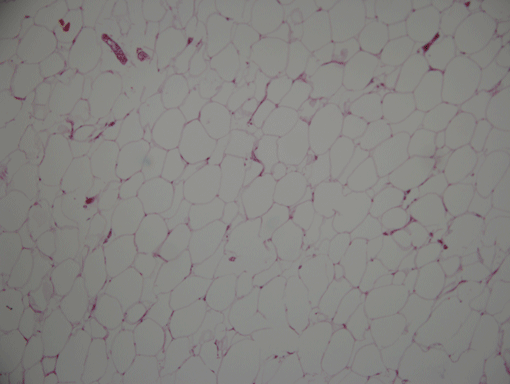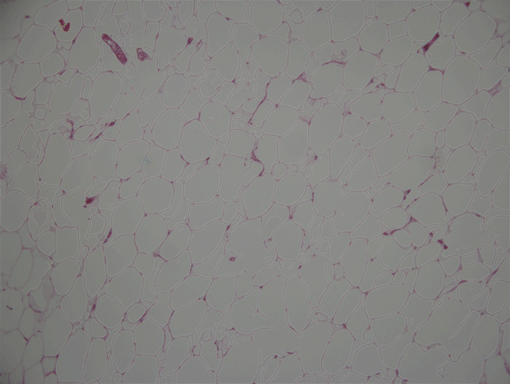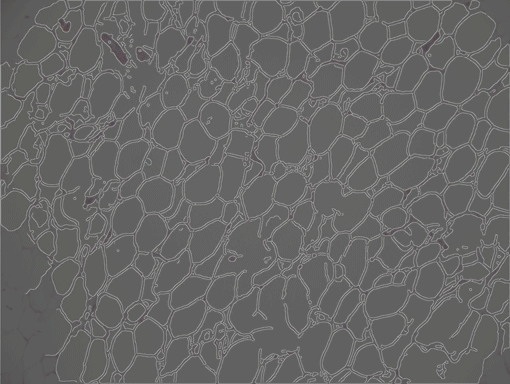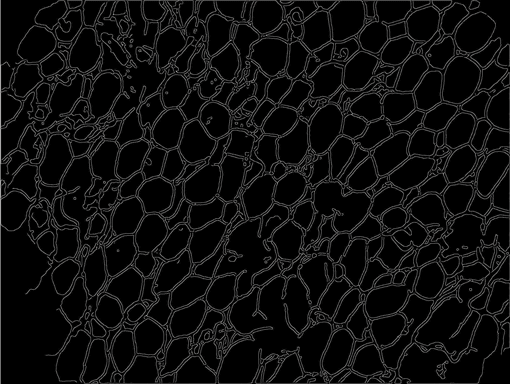
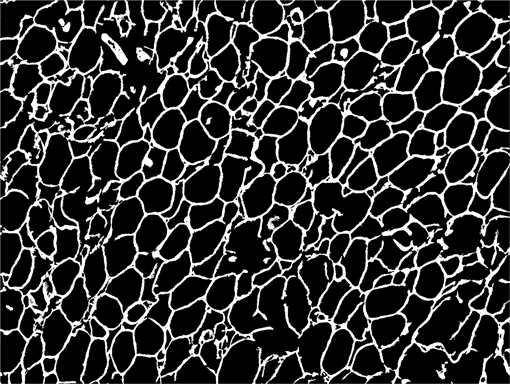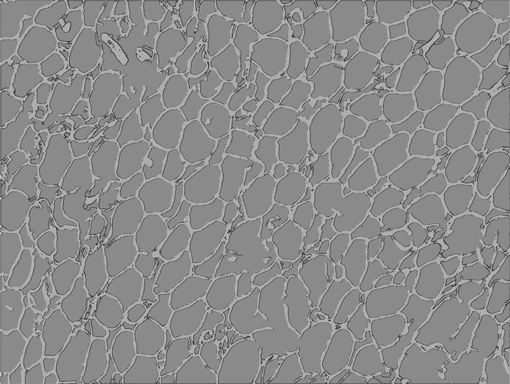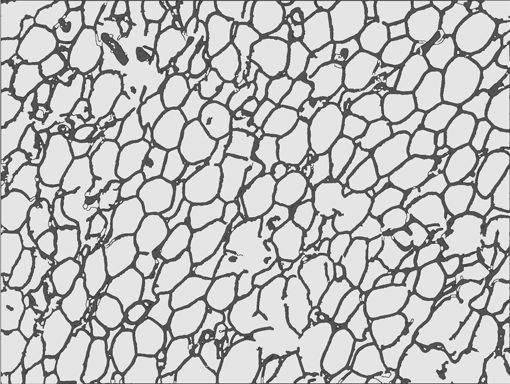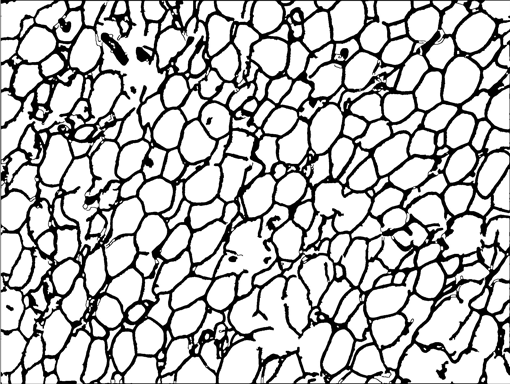
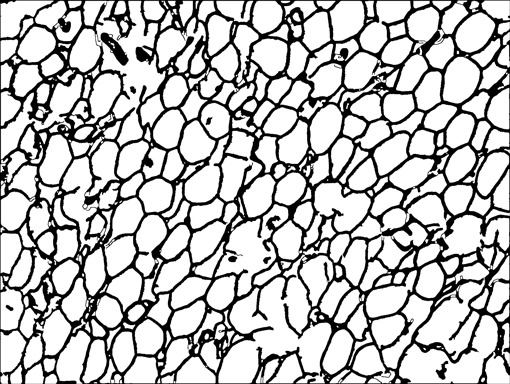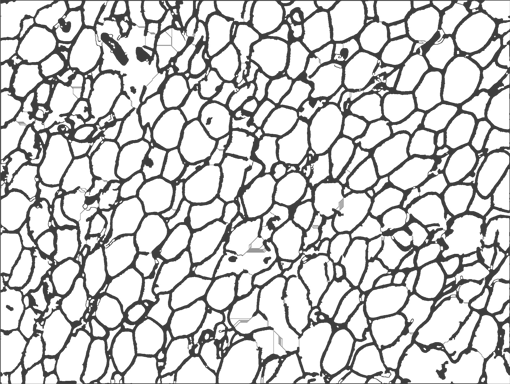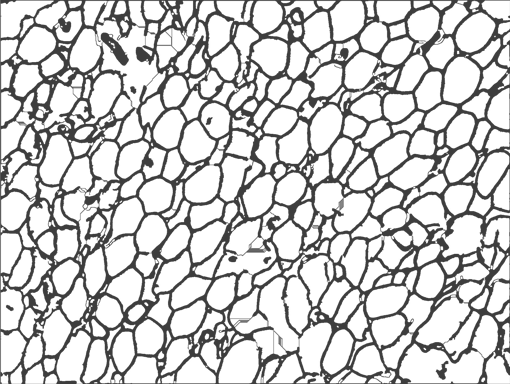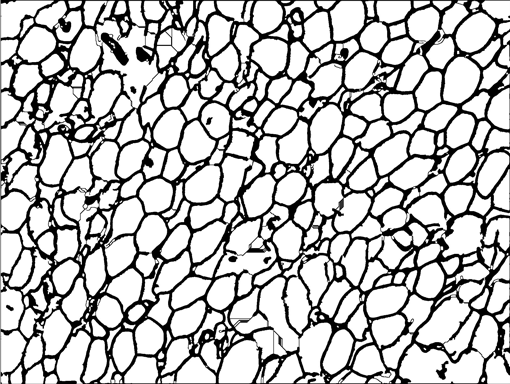
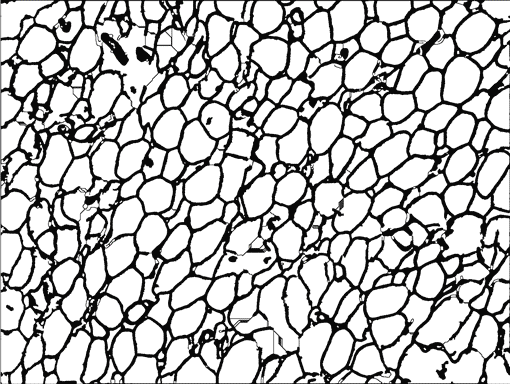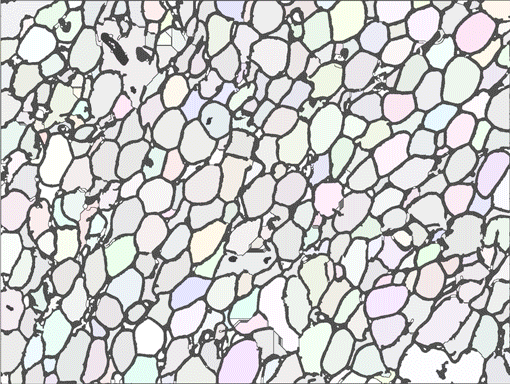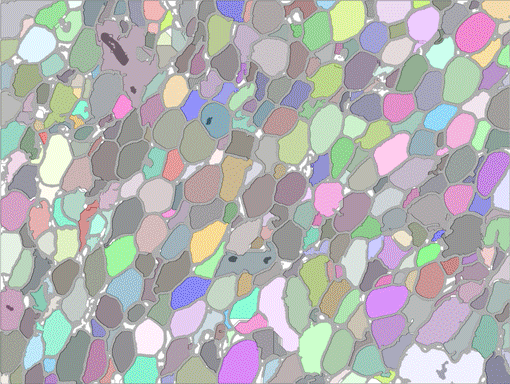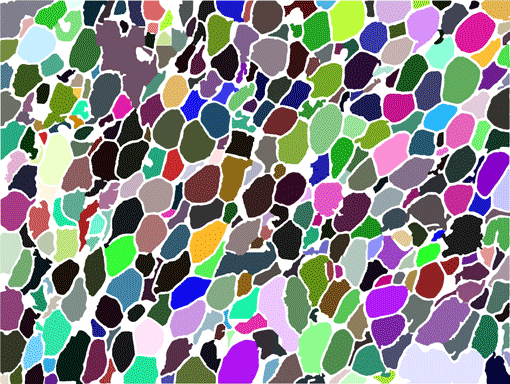
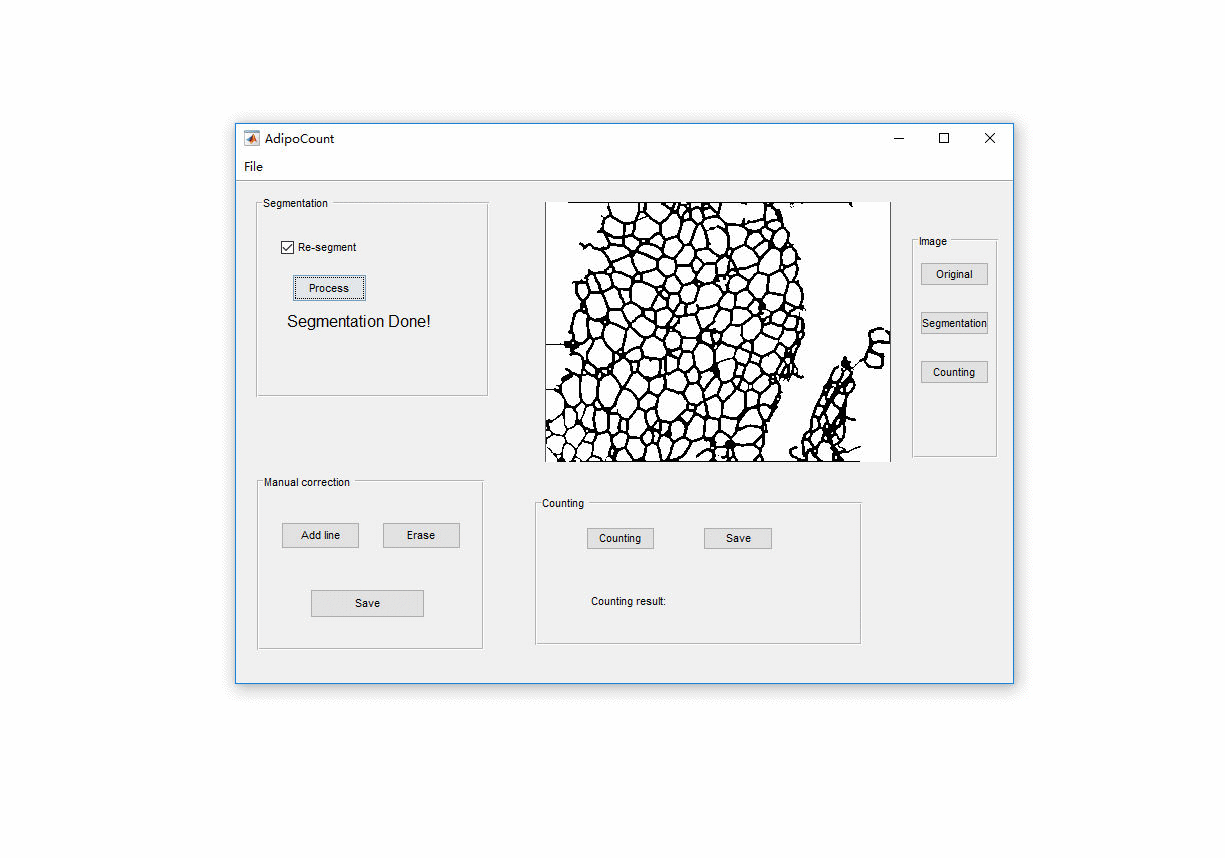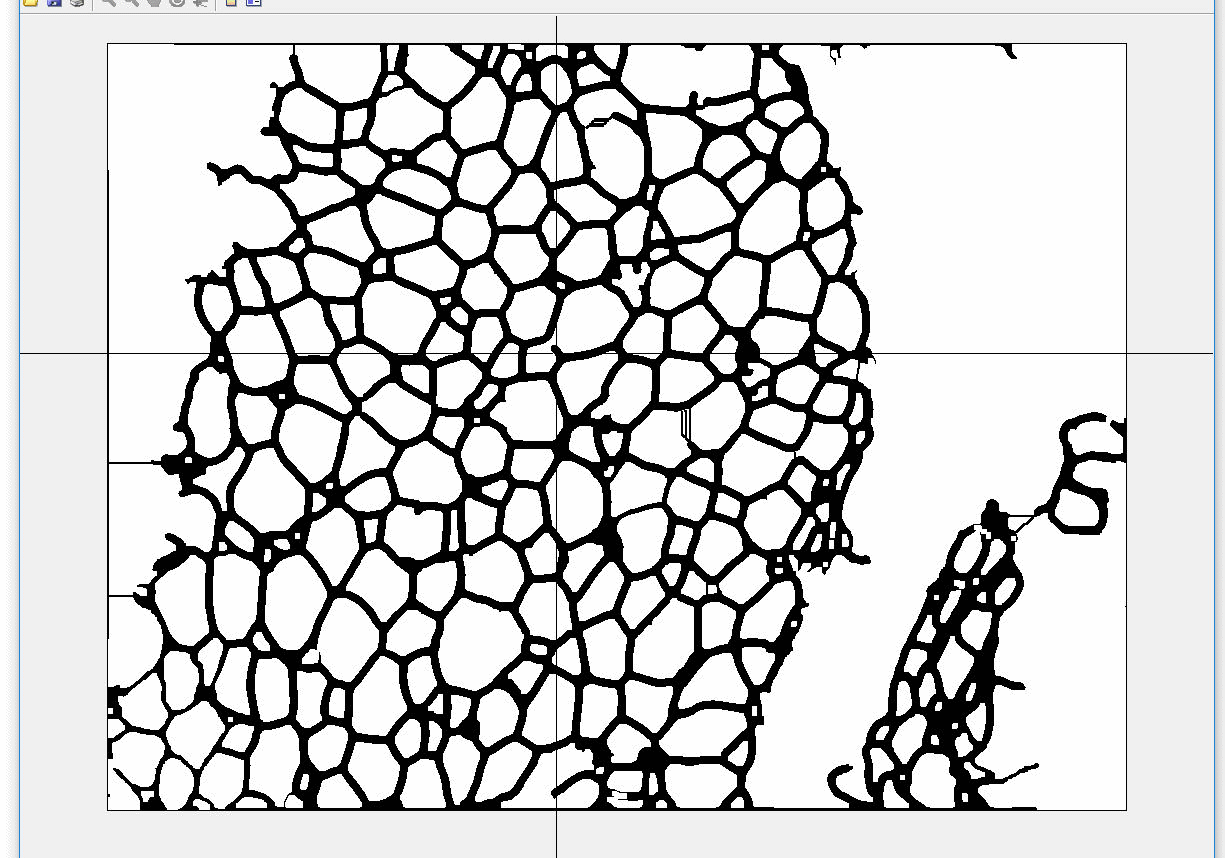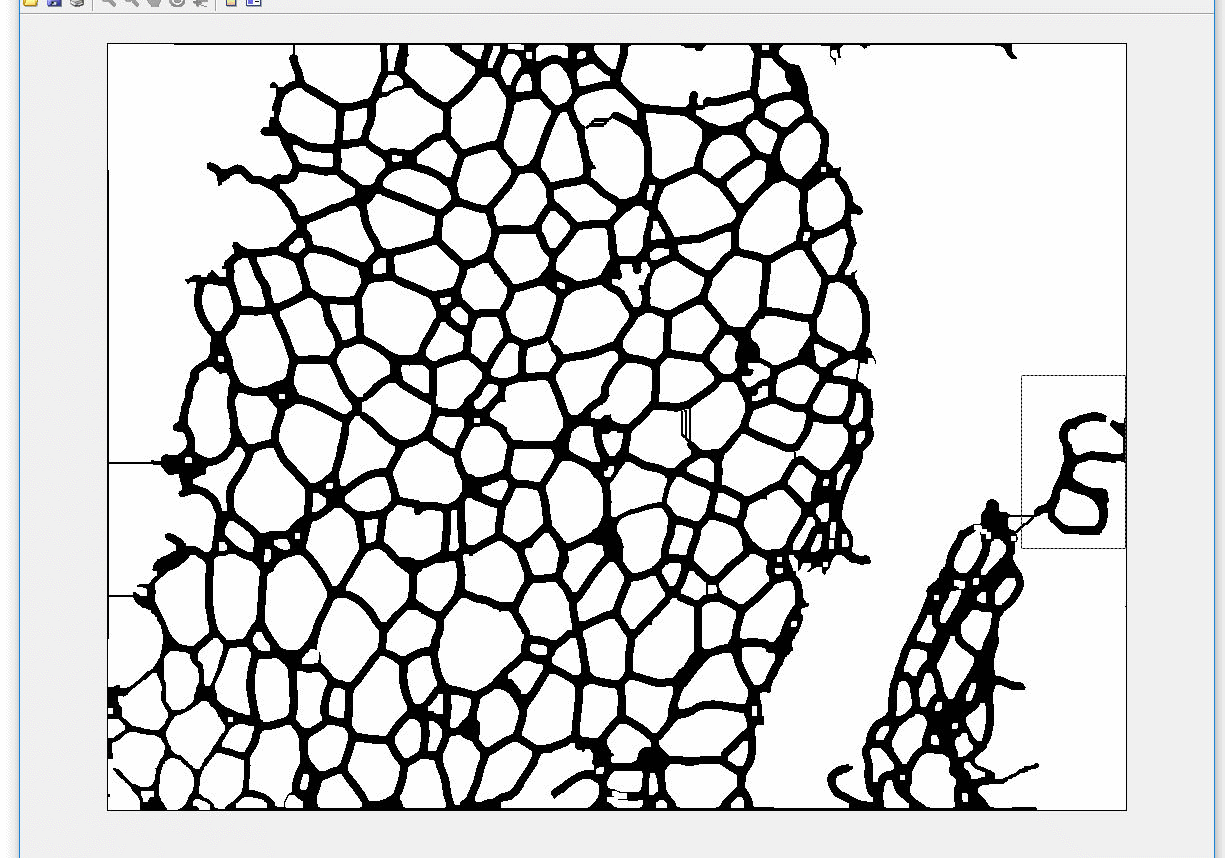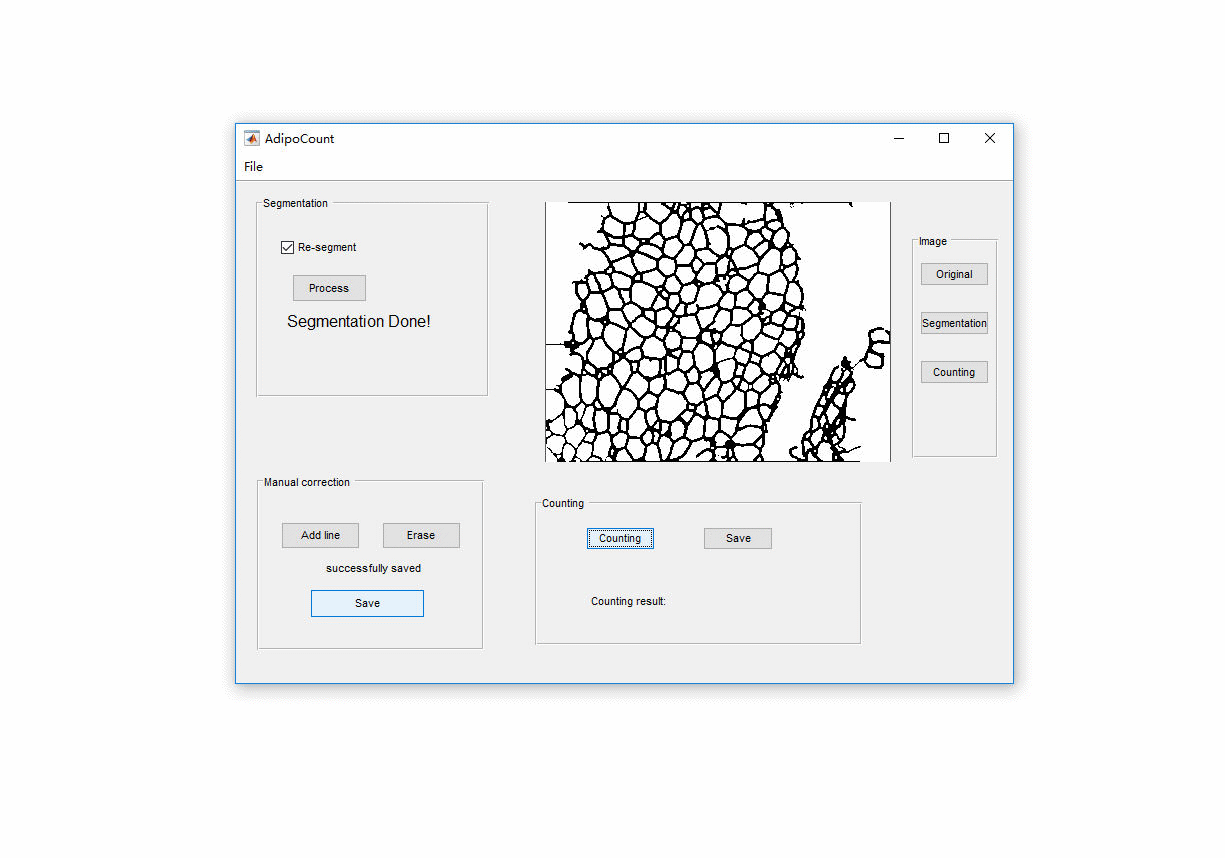
AdipoCount is an automatic adipoytes counting system using image processing algorithms, which can significantly imporve the efficency of adipoytes counting. It contains three modules: membrane segmentation module, re-segmentation module and cell counting module. Membrane segmentation module can segment the membrane, re-segmentation module can estimate some miss-stained membrane and improve the segmentation result, and the cell counting module can detect, count and label them on the image. AdipoCount can generate reliable counting result, which can be further corrected manually.