|
Help of OSML |
| กก |
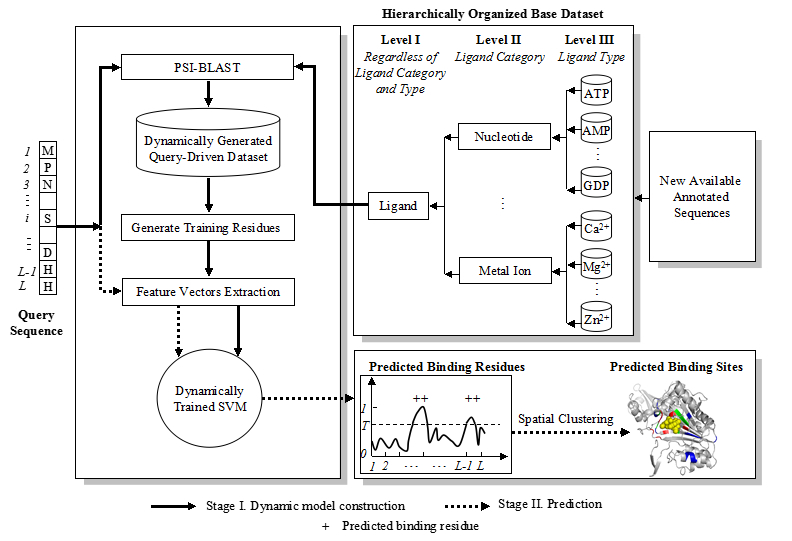
Figure 1. Flowchart of OSML: an on-site implemented predictor for protein-ligand binding sites prediction. Note that in OSML, the base dataset consisting of annotated ligand-binding protein sequences is hierarchically organized to facilitate the user to perform protein-ligand binding sites predictions on three different levels (refer to Fig. 1). More specifically, OSML provides three options, i.e., Level I (Regardless of Ligand Category and Type), Level II (Ligand Category), and Level III (Ligand Type), for user to designate on which level the query-driven dataset will be generated. For the convenience of the subsequent description, we call it a Level I Test if the query-driven dataset is generated on Level I of the base dataset. Similarly, Level II Test and Level III Test denote that the query-driven datasets are generated on Level II and III of the base dataset, respectively. More specifically, Level I Test, Level II Test, and Level III Test are defined as follows: |